Search Count: 11
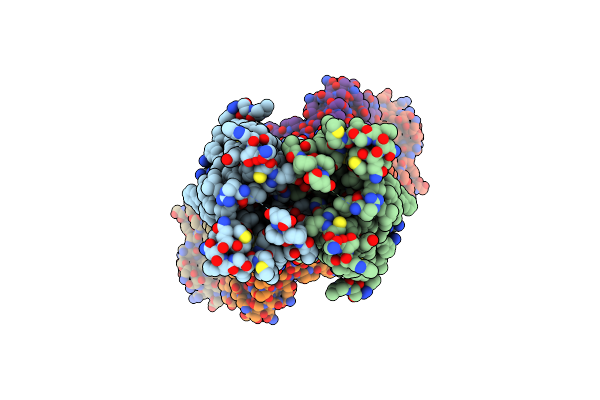 |
Cryo-Em Structure Of Hznt7-Fab Complex In Zinc-Unbound State, Determined In Outward-Facing Conformation
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-09-20 Classification: METAL TRANSPORT |
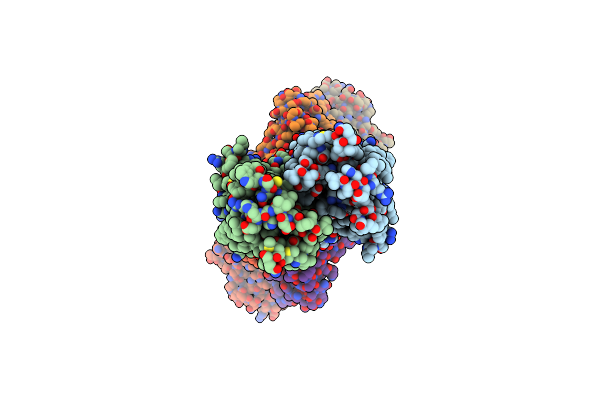 |
Cryo-Em Structure Of Hznt7-Fab Complex In Zinc-Bound State, Determined In Outward-Facing Conformation
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-09-20 Classification: METAL TRANSPORT Ligands: ZN |
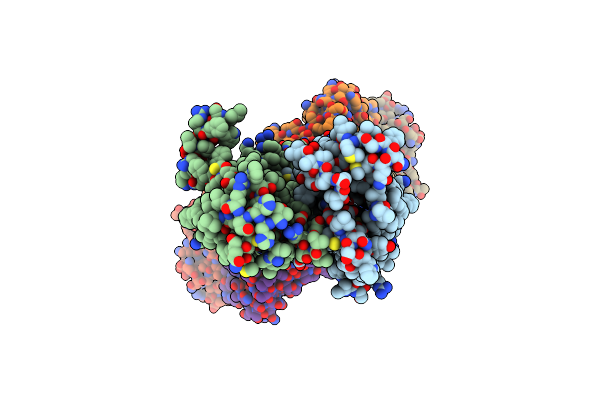 |
Cryo-Em Structure Of Hznt7-Fab Complex In Zinc-Unbound State, Determined In Heterogeneous Conformations- One Subunit In An Inward-Facing And The Other In An Outward-Facing Conformation
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-09-20 Classification: METAL TRANSPORT |
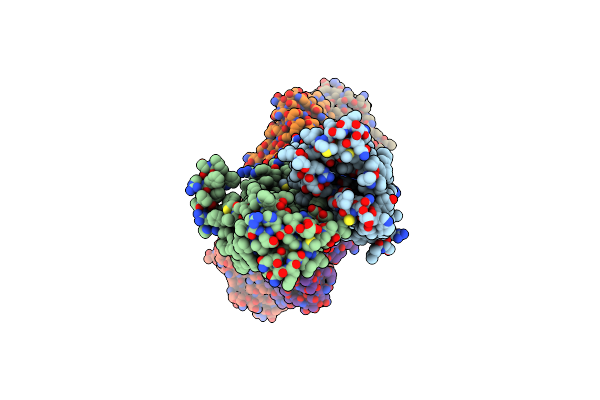 |
Cryo-Em Structure Of Hznt7-Fab Complex In Zinc State 2, Determined In Heterogeneous Conformations- One Subunit In An Inward-Facing Zinc-Bound And The Other In An Outward-Facing Zinc-Bound Conformation
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-09-20 Classification: METAL TRANSPORT Ligands: ZN |
 |
Cryo-Em Structure Of Hznt7Deltahis-Loop-Fab Complex In Zinc-Unbound State, Determined In Outward-Facing Conformation
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-09-20 Classification: METAL TRANSPORT |
 |
Cryo-Em Structure Of Hznt7Deltahis-Loop-Fab Complex In Zinc-Bound State, Determined In Outward-Facing Conformation
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-09-20 Classification: METAL TRANSPORT Ligands: ZN |
 |
Cryo-Em Structure Of Hznt7-Fab Complex In Zinc State 1, Determined In Heterogeneous Conformations- One Subunit In An Inward-Facing Zinc-Bound And The Other In An Outward-Facing Zinc-Unbound Conformation
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-09-20 Classification: METAL TRANSPORT Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2019-03-13 Classification: CELL CYCLE Ligands: EPE |
 |
Organism: Klebsiella oxytoca
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-02-22 Classification: LYASE Ligands: GOL, CA, PO4, B12 |
 |
 |
Crystal Structure Of The Conserved Protein Tt1542 From Thermus Thermophilus Hb8
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2003-08-05 Classification: structural genomics, unknown function |

