Search Count: 35
 |
Organism: Measles morbillivirus
Method: SOLUTION NMR Release Date: 2025-09-03 Classification: VIRUS Ligands: ZN |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN |
 |
Organism: Synthetic construct, Escherichia phage t7
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2023-02-01 Classification: Transferase/DNA/RNA Ligands: GOL |
 |
Organism: Synthetic construct, Escherichia phage t7
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2023-02-01 Classification: Transferase/DNA/RNA Ligands: GOL, S8L, MG |
 |
Organism: Synthetic construct, Escherichia phage t7
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2023-02-01 Classification: Transferase/DNA/RNA Ligands: GOL, 3PO, MG |
 |
Organism: Synthetic construct, Escherichia phage t7
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2023-02-01 Classification: Transferase/DNA/RNA |
 |
Organism: Synthetic construct, Escherichia phage t7
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-02-01 Classification: Transferase/DNA/RNA Ligands: S96, MG |
 |
Organism: Synthetic construct, Escherichia phage t7
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2023-02-01 Classification: Transferase/DNA/RNA Ligands: ATP, MG |
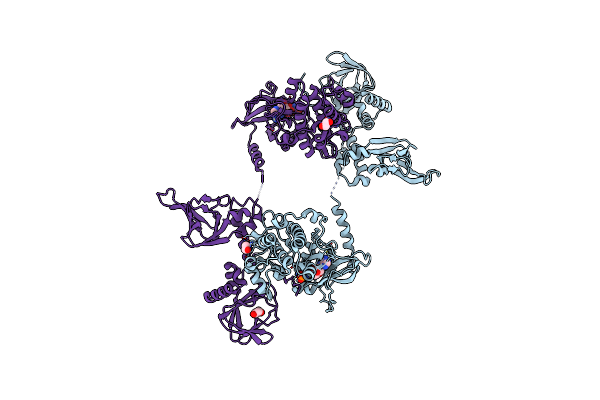 |
Crystal Structure Of Human Protein Kinase G (Pkg) R-C Complex In Inhibited State
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2022-08-24 Classification: TRANSFERASE/INHIBITOR Ligands: MN, ANP, EDO |
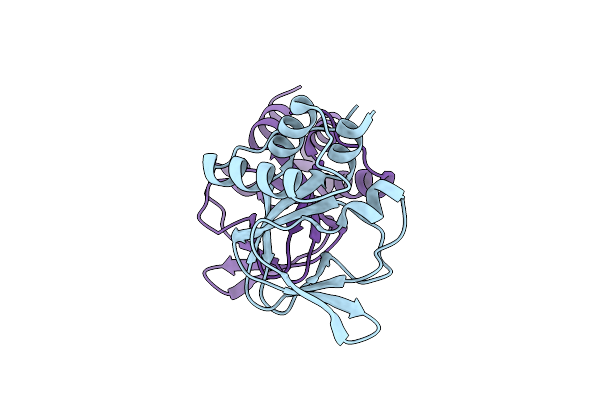 |
Crystal Structure Of Cgmp Dependent Protein Kinase I Alpha (Pkg I Alpha)Cnb-A Domain With R177Q Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.26 Å Release Date: 2022-05-11 Classification: SIGNALING PROTEIN |
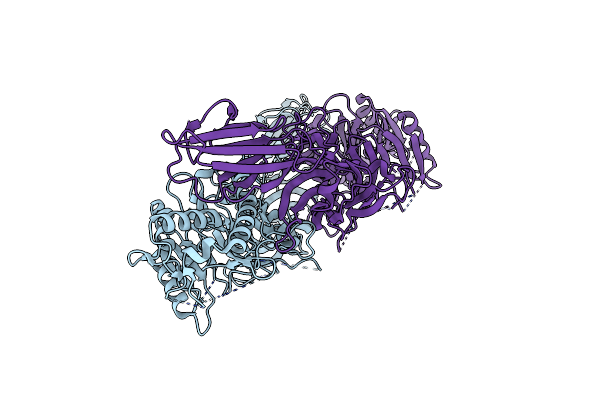 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.96 Å Release Date: 2021-06-02 Classification: HYDROLASE |
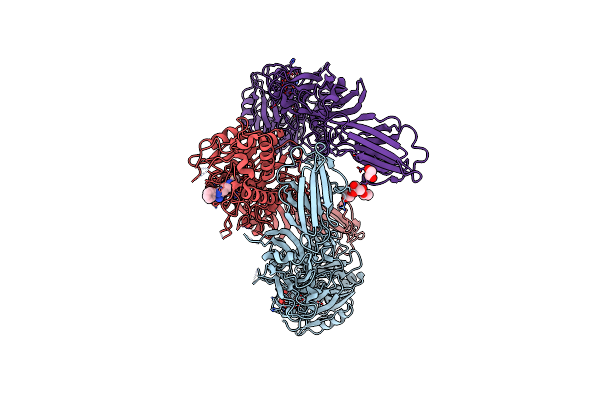 |
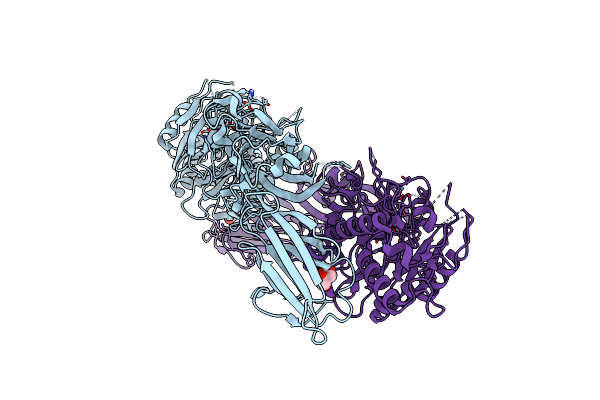
Structure Of The Peptidylarginine Deiminase Type Iii (Pad3) In Complex With Cl-Amidine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.18 Å Release Date: 2021-06-02 Classification: HYDROLASE Ligands: BFB, CA, CL, EDO, GOL |
 |
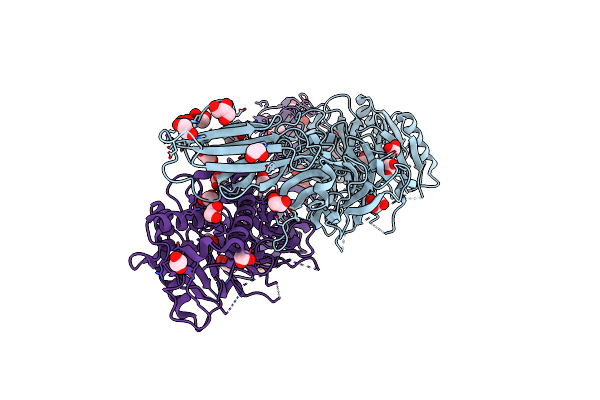
Structure Of The Ca2+-Bound C646A Mutant Of Peptidylarginine Deiminase Type Iii (Pad3)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2021-06-02 Classification: HYDROLASE Ligands: CA, GOL, CL |
 |
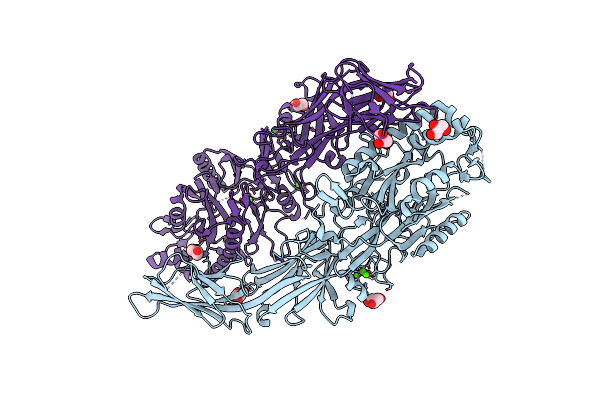
Structure Of The C646A Mutant Of Peptidylarginine Deiminase Type Iii (Pad3)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-06-02 Classification: HYDROLASE Ligands: EDO, GOL |
 |
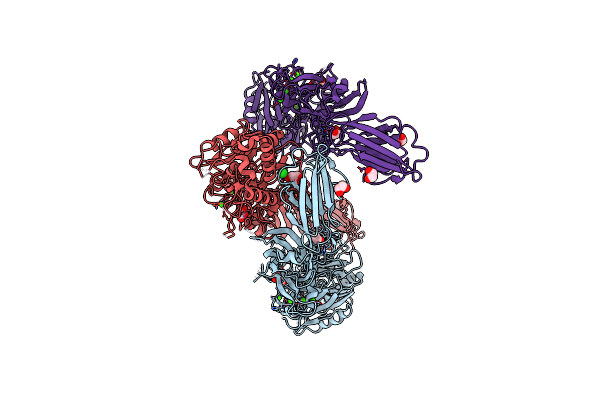
Structure Of The Inactive Form Of Wild-Type Peptidylarginine Deiminase Type Iii (Pad3) Crystallized Under The Condition With High Concentrations Of Ca2+
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2021-06-02 Classification: HYDROLASE Ligands: CA, GOL, CL |
 |
Structure Of The Ca2+-Bound Wild-Type Peptidylarginine Deiminase Type Iii (Pad3)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2021-06-02 Classification: HYDROLASE Ligands: CA, CL, EDO, GOL |
 |
Crystal Structure Of A211D Mutant Of Protein Kinase A Ria Subunit, A Carney Complex Mutation
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:4.16 Å Release Date: 2021-05-26 Classification: TRANSFERASE Ligands: CMP |
 |
Organism: Anthopleura elegantissima
Method: SOLUTION NMR Release Date: 2020-12-23 Classification: TOXIN |
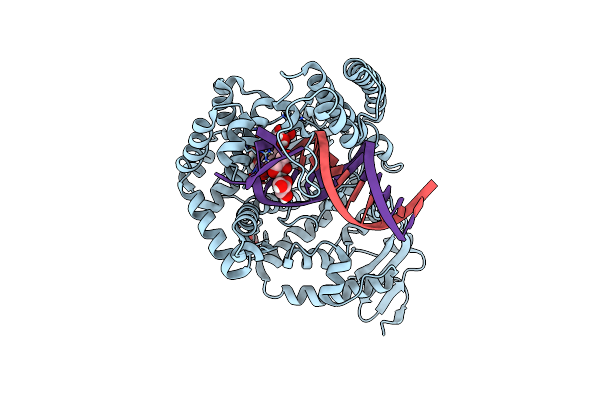 |
Crystal Structure Of The Large Fragment Of Dna Polymerase I From Thermus Aquaticus In A Closed Ternary Complex With The Artificial Base Pair Dds-Dpxtp
Organism: Thermus aquaticus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-06-28 Classification: TRANSFERASE Ligands: MG, ACT, 91T, 91R, GOL |
 |
Organism: Pyrococcus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-01-25 Classification: TRANSFERASE |

