Search Count: 80
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: IMMUNE SYSTEM Ligands: GAL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: IMMUNE SYSTEM Ligands: FUL |
 |
Cryo-Em Structure Of The Potassium-Selective Channelrhodopsin Hckcr1 In Lipid Nanodisc
Organism: Hyphochytrium catenoides
Method: ELECTRON MICROSCOPY Resolution:2.56 Å Release Date: 2023-09-06 Classification: MEMBRANE PROTEIN Ligands: RET, PSC, PLM |
 |
Cryo-Em Structure Of The Potassium-Selective Channelrhodopsin Hckcr2 In Lipid Nanodisc
Organism: Hyphochytrium catenoides
Method: ELECTRON MICROSCOPY Resolution:2.53 Å Release Date: 2023-09-06 Classification: MEMBRANE PROTEIN Ligands: RET, PSC, PLM |
 |
Cryo-Em Structure Of The Potassium-Selective Channelrhodopsin Hckcr1 H225F Mutant In Lipid Nanodisc
Organism: Hyphochytrium catenoides
Method: ELECTRON MICROSCOPY Resolution:2.66 Å Release Date: 2023-09-06 Classification: MEMBRANE PROTEIN Ligands: RET, PSC, PLM |
 |
Organism: Staphylococcus aureus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2023-07-26 Classification: PROTEIN BINDING Ligands: SO4, MLI |
 |
Organism: Staphylococcus aureus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2023-07-26 Classification: PROTEIN BINDING Ligands: SO4 |
 |
Organism: Staphylococcus aureus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2023-07-26 Classification: PROTEIN BINDING Ligands: GOL |
 |
Organism: Staphylococcus aureus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-07-26 Classification: PROTEIN BINDING |
 |
Organism: Staphylococcus aureus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.02 Å Release Date: 2023-07-26 Classification: PROTEIN BINDING Ligands: MLI |
 |
Organism: Staphylococcus aureus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2023-07-26 Classification: PROTEIN BINDING Ligands: GOL |
 |
Organism: Staphylococcus aureus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2023-07-26 Classification: PROTEIN BINDING Ligands: GOL, SO4 |
 |
Organism: Staphylococcus aureus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2023-07-26 Classification: PROTEIN BINDING Ligands: MLI |
 |
Organism: Staphylococcus aureus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2023-07-26 Classification: PROTEIN BINDING |
 |
Organism: Staphylococcus aureus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2023-07-26 Classification: PROTEIN BINDING Ligands: GOL |
 |
Organism: Rhodomonas lens
Method: ELECTRON MICROSCOPY Resolution:2.02 Å Release Date: 2022-02-02 Classification: MEMBRANE PROTEIN Ligands: RET, CLR, PLM |
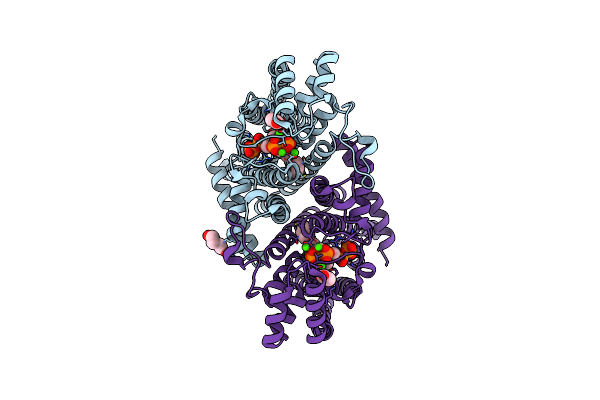 |
Organism: Leishmania major
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-11-25 Classification: TRANSFERASE Ligands: CA, IPR, 476, ACT, PEG |
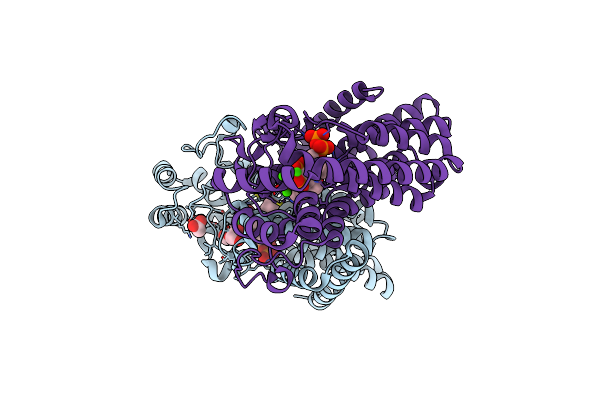 |
Organism: Leishmania major
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-10-07 Classification: TRANSFERASE Ligands: CA, ACT, IPR, 476, PEG |
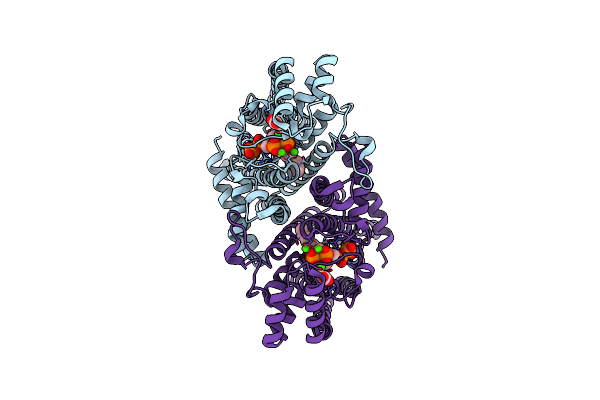 |
Organism: Leishmania major
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2020-10-07 Classification: TRANSFERASE Ligands: CA, IPR, 476, ACT |
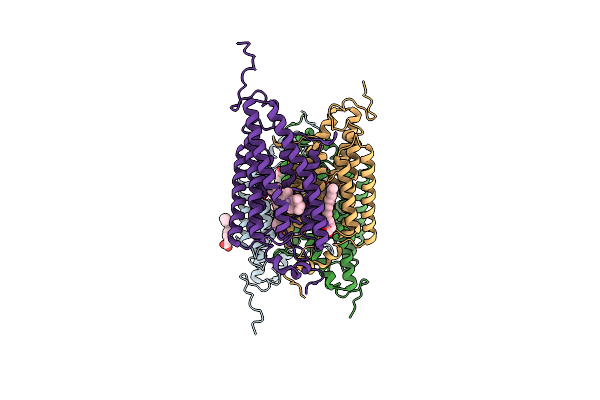 |
Organism: Guillardia theta ccmp2712
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2018-09-05 Classification: MEMBRANE PROTEIN Ligands: RET, OLA |

