Search Count: 48
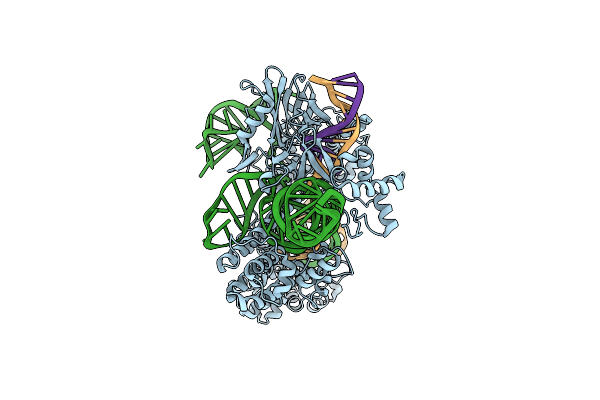 |
Cryo-Em Structure Of Small And Dead Form Sacas9-Rna-Dna Ternary Complex (Sdcas9)
Organism: Staphylococcus aureus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-04-02 Classification: DNA BINDING PROTEIN |
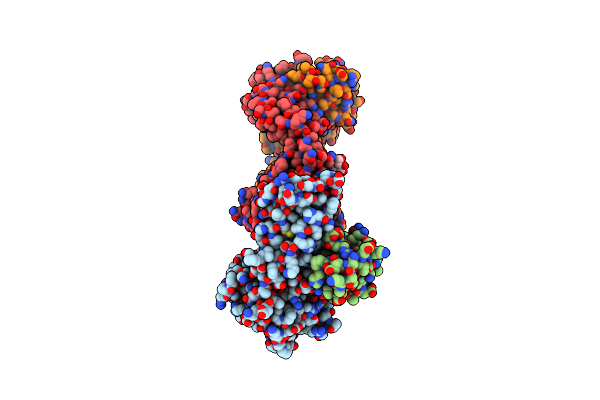 |
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-02-05 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
Crystal Structure Of Co Dehydrogenase Mutant With Increased Affinity For Electron Mediators In High Peg Concentration
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2024-04-17 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, XCC, FE, 1PE |
 |
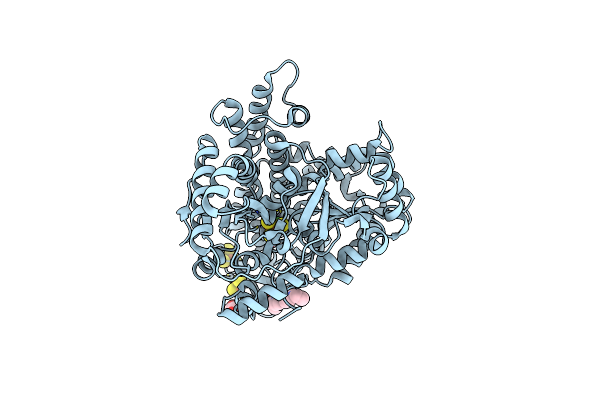
Crystal Structure Of Co Dehydrogenase Mutant With Increased Affinity For Electron Mediators In Low Peg Concentration
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-04-17 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, XCC, FE, EDO |
 |
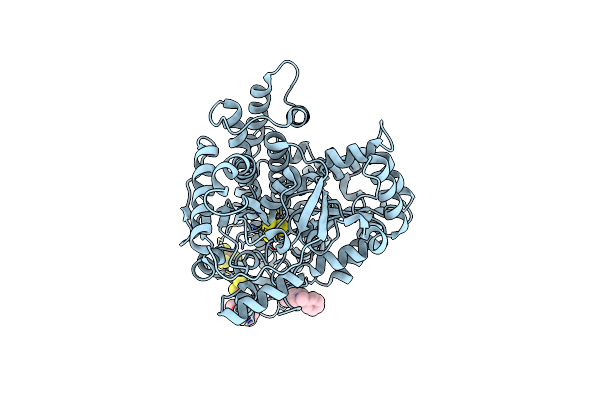
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2024-04-17 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, XCC, S8I, EDO |
 |
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2024-04-17 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, XCC, PGE, S7L |
 |
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-04-17 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, XCC, FE |
 |
Organism: Methylorubrum extorquens am1
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: ELECTRON TRANSPORT Ligands: W, MGD, FES, SF4, FMN |
 |
Organism: Carboxydothermus hydrogenoformans
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2022-10-19 Classification: OXIDOREDUCTASE Ligands: SF4, FES, NFS |
 |
Tunnel-Redesigned O2-Tolerant Co Dehydrogenase For Removal Of Co In Real Flue Gas (Chcodh2 A559H Mutant In Anaerobic Condition)
Organism: Carboxydothermus hydrogenoformans
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-10-19 Classification: OXIDOREDUCTASE Ligands: SF4, FES, NFS |
 |
Tunnel-Redesigned O2-Tolerant Co Dehydrogenase For Removal Of Co In Real Flue Gas (Chcodh2 A559S Mutant In Anaerobic Condition)
Organism: Carboxydothermus hydrogenoformans
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2022-10-19 Classification: OXIDOREDUCTASE Ligands: SF4, FES, NFS |
 |
Tunnel-Redesigned O2-Tolerant Co Dehydrogenase For Removal Of Co In Real Flue Gas (Aerobic Chcodh2 A559W Mutant)
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2022-10-05 Classification: OXIDOREDUCTASE Ligands: SF4, FES, NFS |
 |
Organism: Bacillus velezensis fzb42
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-01-19 Classification: PROTEIN BINDING Ligands: CL |
 |
Plant Growth-Promoting Factor Yxal Mutant From Bacillus Velezensis - T175W/W215G
Organism: Bacillus velezensis fzb42
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-01-12 Classification: SIGNALING PROTEIN |
 |
Plant Growth-Promoting Factor Yxal Mutant From Bacillus Velezensis - T175W/S213G/W215A
Organism: Bacillus velezensis (strain dsm 23117 / bgsc 10a6 / fzb42)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-01-12 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-12-22 Classification: TRANSFERASE Ligands: G5R, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2021-06-23 Classification: METAL BINDING PROTEIN Ligands: CA, CL, SCN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2021-06-23 Classification: METAL BINDING PROTEIN Ligands: CA, ACT |
 |
Human Calcium And Integrin Binding Protein 3 Bound To Tmc1 Residues 303-347
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2021-06-23 Classification: METAL BINDING/TRANSPORT PROTEIN Ligands: MG |
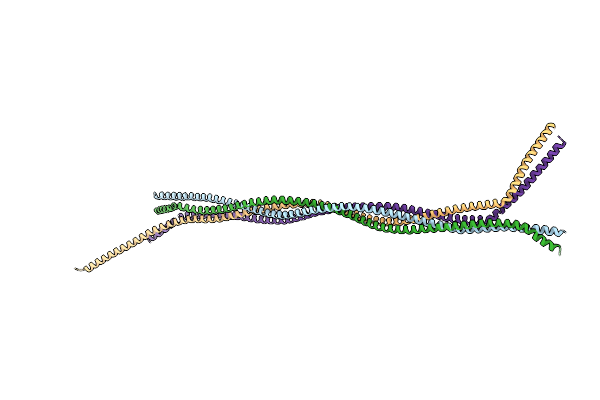 |
Crystal Structure Of Lamin A/C Fragment And Assembly Mechanisms Of Intermediate Filaments
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.21 Å Release Date: 2019-09-11 Classification: STRUCTURAL PROTEIN |

