Search Count: 23
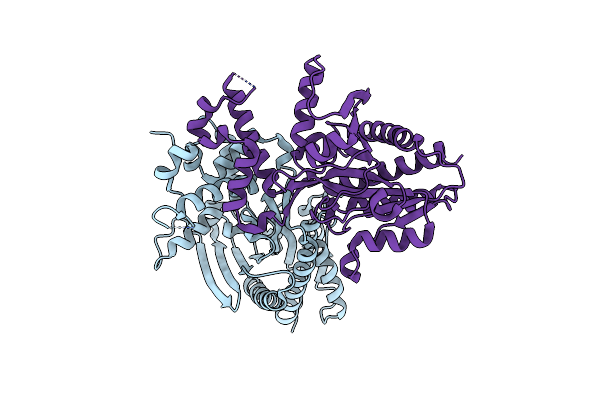 |
Crystal Structure Of Germination Protease From The Spore-Forming Bacterium Paenisporosarcina Sp. Tg-20 In Its Inactive Form
Organism: Paenisporosarcina sp.
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-03-31 Classification: PROTEASE |
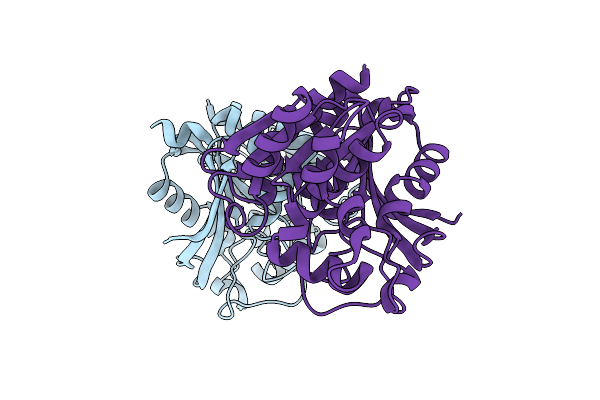 |
S-Formylglutathione Hydrolase Homolog From A Psychrophilic Bacterium Of Shewanella Frigidimarina
Organism: Shewanella frigidimarina
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2019-09-04 Classification: HYDROLASE |
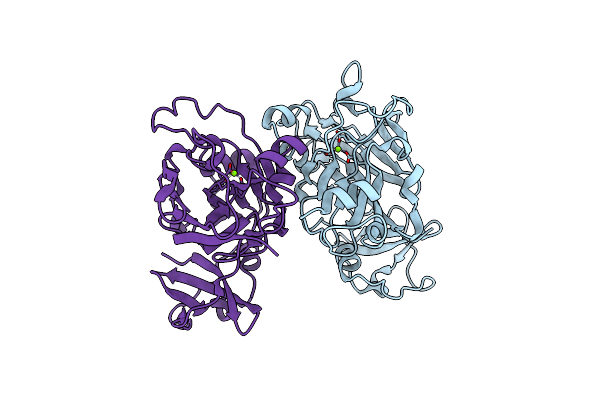 |
Fumarylacetoacetate Hydrolase (Eafah) From Psychrophilic Exiguobacterium Antarcticum
Organism: Exiguobacterium antarcticum b7
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-04-03 Classification: HYDROLASE Ligands: MG |
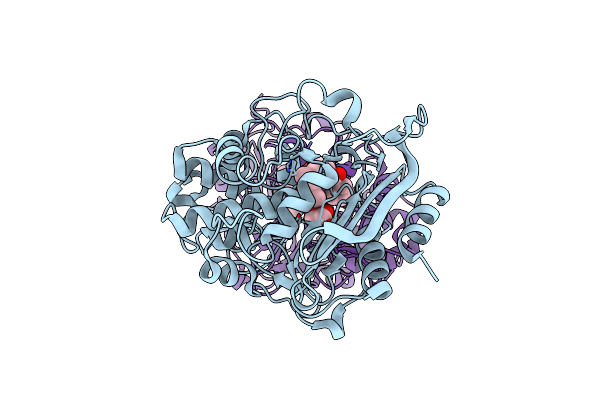 |
Structural Basis For The Enantioselectivity Of Est-Y29 Toward (S)-Ketoprofen
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-03-13 Classification: HYDROLASE Ligands: GOL, 9KF |
 |
Structural Basis For The Enantioselectivity Of Est-Y29 Toward (S)-Ketoprofen
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2019-03-13 Classification: HYDROLASE Ligands: GOL, 9KL |
 |
Structural Basis For The Enantioselectivity Of Est-Y29 Toward (S)-Ketoprofen
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-03-13 Classification: HYDROLASE |
 |
Organism: Paenibacillus sp. r4
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-10-10 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of A Novel Penicillin-Binding Protein (Pbp) Homolog From Caulobacter Crescentus
Organism: Caulobacter crescentus (strain atcc 19089 / cb15)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-07-12 Classification: HYDROLASE |
 |
Organism: Exiguobacterium antarcticum (strain b7)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-01-11 Classification: HYDROLASE Ligands: F50 |
 |
Crystal Structrue Of Est24, A Carbohydrate Acetylesterase From Sinorhizobium Meliloti
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2016-04-27 Classification: HYDROLASE Ligands: PO4, P6G |
 |
The Crystal Structure Of Carbohydrate Acetylesterase Family Member From Sinorhizobium Meliloti
Organism: Sinorhizobium meliloti 1021
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2014-12-24 Classification: HYDROLASE |
 |
Crystal Structure Of Est-Y29,A Novel Penicillin-Binding Protein/Beta-Lactamase Homolog From A Metagenomic Library
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-09-10 Classification: HYDROLASE |
 |
Crystal Structure Of Est-Y29, A Novel Penicillin-Binding Protein/Beta-Lactamase Homolog From A Metagenomic Library
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-09-10 Classification: HYDROLASE Ligands: DEP |
 |
Crystal Structure Of Est-Y29, A Novel Penicillin-Binding Protein/Beta-Lactamase Homolog From A Metagenomic Library
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-09-10 Classification: HYDROLASE Ligands: 4NP |
 |
Structural Plasticity In Igsf Domain 4 Of Icam-1 Mediates Cell Surface Dimerization
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2007-10-16 Classification: CELL ADHESION Ligands: NAG, SO4, ZN, TRS |
 |
Structural Basis Of Non-Specific Lipid Binding In Maize Lipid-Transfer Protein Complexes With Capric Acid Revealed By High-Resolution X-Ray Crystallography
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2001-06-06 Classification: LIPID TRANSPORT Ligands: DKA, FMT |
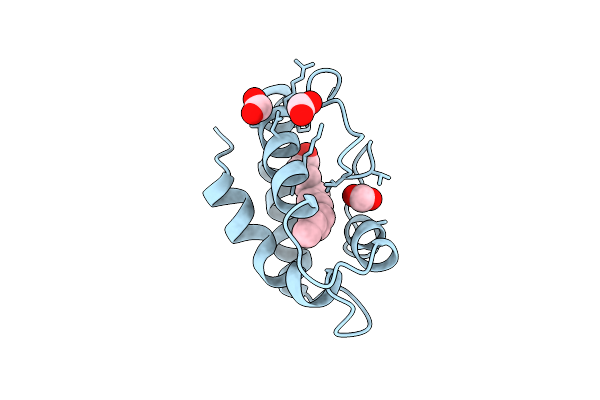 |
Structural Basis Of Non-Specific Lipid Binding In Maize Lipid-Transfer Protein Complexes With Lauric Acid Revealed By High-Resolution X-Ray Crystallography
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2001-06-06 Classification: LIPID TRANSPORT Ligands: DAO, FMT |
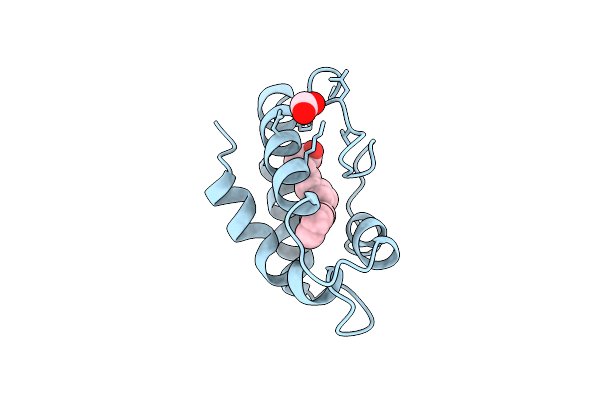 |
Structural Basis Of Non-Specific Lipid Binding In Maize Lipid-Transfer Protein Complexes With Myristic Acid Revealed By High-Resolution X-Ray Crystallography
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2001-06-06 Classification: LIPID TRANSPORT Ligands: MYR, FMT |
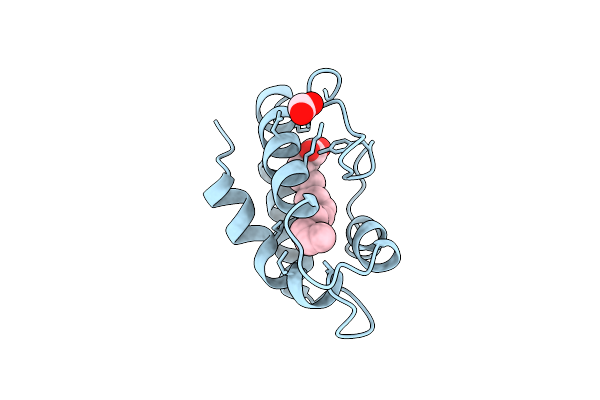 |
Structural Basis Of Non-Specific Lipid Binding In Maize Lipid-Transfer Protein Complexes With Palmitoleic Acid Revealed By High-Resolution X-Ray Crystallography
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2001-06-06 Classification: LIPID TRANSPORT Ligands: PAM, FMT |
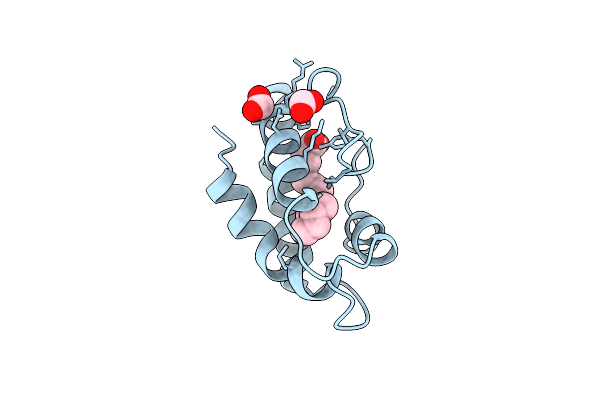 |
Structural Basis Of Non-Specific Lipid Binding In Maize Lipid-Transfer Protein Complexes With Stearic Acid Revealed By High-Resolution X-Ray Crystallography
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2001-06-06 Classification: LIPID TRANSPORT Ligands: STE, FMT |

