Search Count: 34
 |
Crystal Structure Of Iron-Bound Human Ado C18S/C239S Variant Soaked In Hydralazine At 2.39 Angstrom Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: OXIDOREDUCTASE Ligands: FE2, HLZ, GOL |
 |
Crystal Structure Of Cobalt-Bound Human Ado C18S/C239S Variant In Complex With Hydralazine At 1.89 Angstrom Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: OXIDOREDUCTASE Ligands: CO, HLZ, GOL, SO4 |
 |
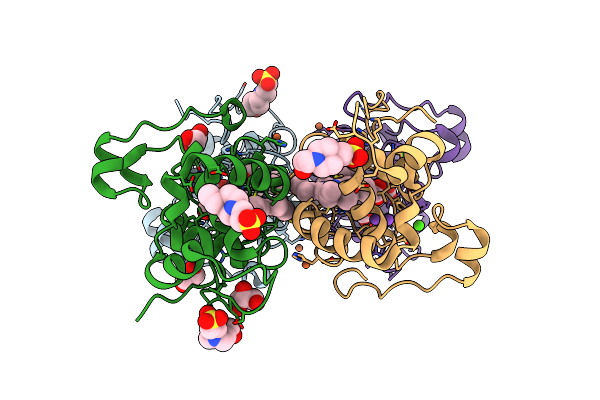
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-06-21 Classification: VIRAL PROTEIN/IMMUNE SYSTEM/DNA Ligands: NAG |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-06-21 Classification: VIRAL PROTEIN/IMMUNE SYSTEM/DNA Ligands: NAG |
 |
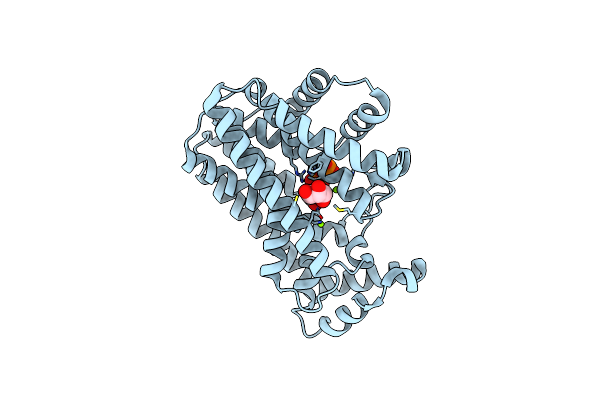
Crystal Structure Of Cytochrome Cl From The Marine Methylotrophic Bacterium Methylophaga Aminisulfidivorans Mpt (Ma-Cytcl)
Organism: Methylophaga aminisulfidivorans mp
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2020-07-22 Classification: ELECTRON TRANSPORT Ligands: HEC, EPE, GOL, CA, FE, NA |
 |
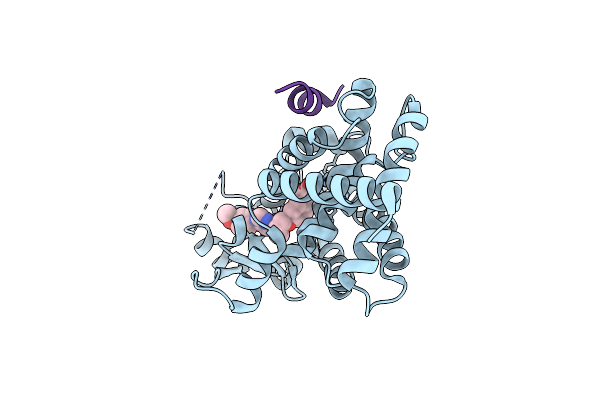
Organism: Nonlabens dokdonensis (strain dsm 17205 / kctc 12402 / dsw-6)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-09-11 Classification: TRANSFERASE Ligands: PPV, GOL, MG |
 |
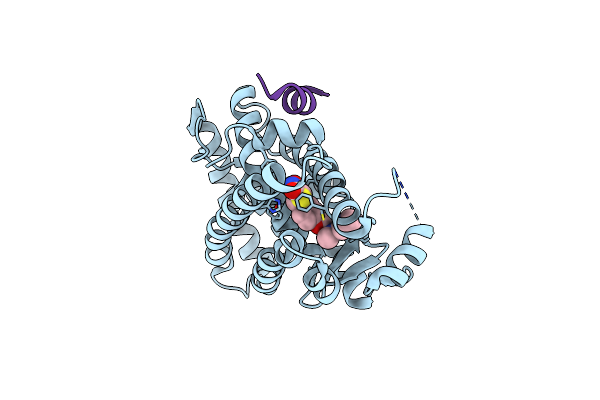
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2018-09-12 Classification: TRANSCRIPTION Ligands: 8LX |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-09-12 Classification: TRANSCRIPTION Ligands: BRL |
 |
Crystal Structure Of Methanol Dehydrogenase From Methylophaga Aminisulfidivorans
Organism: Methylophaga aminisulfidivorans mp
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-03-21 Classification: OXIDOREDUCTASE Ligands: PQQ, MG |
 |
Organism: Methylophaga aminisulfidivorans mp
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2017-07-12 Classification: UNKNOWN FUNCTION Ligands: BR |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2015-10-21 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 4MG |
 |
Crystal Structure Of The Mevalonate Kinase From An Archaeon Methanosarcina Mazei
Organism: Methanosarcina mazei
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2012-12-12 Classification: TRANSFERASE Ligands: MG |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2011-11-23 Classification: LIGASE Ligands: ZN |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-11-23 Classification: PROTEIN TRANSPORT |
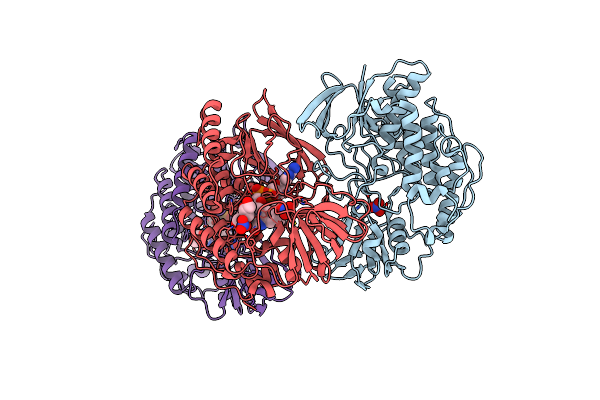 |
Organism: Methylophaga aminisulfidivorans
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2011-05-04 Classification: OXIDOREDUCTASE Ligands: NO3, FAD, GOL |
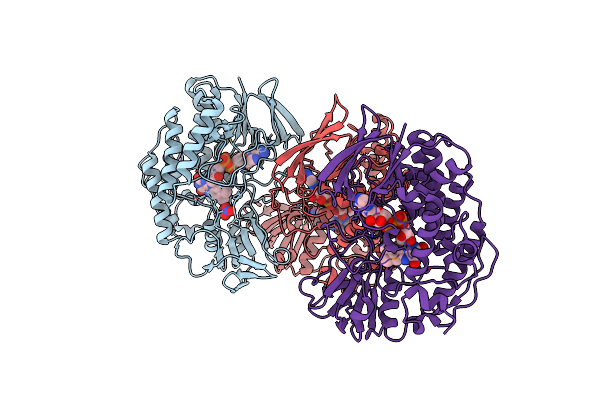 |
Organism: Methylophaga aminisulfidivorans
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2011-05-04 Classification: OXIDOREDUCTASE Ligands: FAD, NO3, GOL |
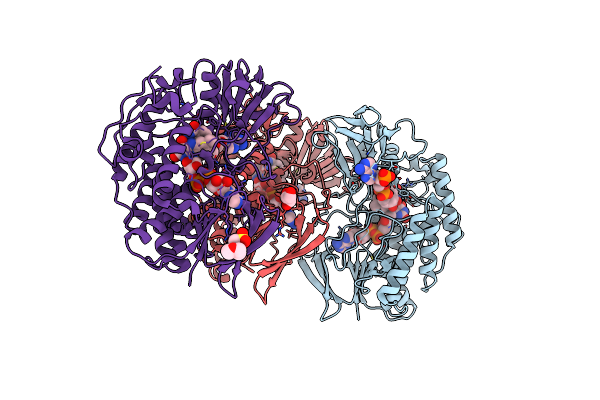 |
Crystal Structure Of Bacterial Flavin Containing Monooxygenase In Complex With Nadp
Organism: Methylophaga aminisulfidivorans
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2011-05-04 Classification: OXIDOREDUCTASE Ligands: FAD, NAP, GOL, MES |
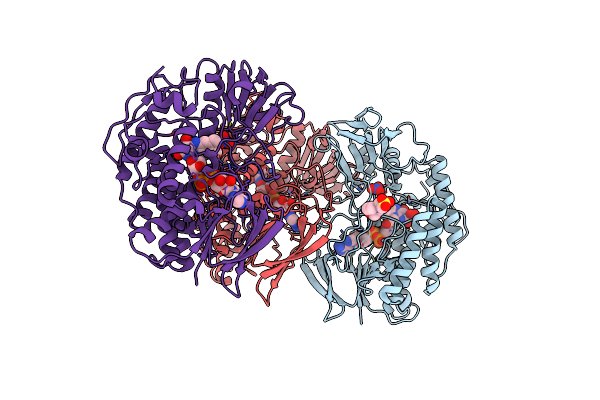 |
Crystal Structure Of The Mutant Bacterial Flavin Containing Monooxygenase (Y207S)
Organism: Methylophaga aminisulfidivorans
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2011-05-04 Classification: OXIDOREDUCTASE Ligands: FAD, NO3, MES, OXY |
 |
Crystal Structure Of The Mutant Bacterial Flavin Containing Monooxygenase In Complex With Indole
Organism: Methylophaga aminisulfidivorans
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2011-05-04 Classification: OXIDOREDUCTASE Ligands: FAD, NO3, IND, OXY, MES |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-11-03 Classification: HYDROLASE Ligands: CL |

