Search Count: 228
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2024-12-11 Classification: HYDROLASE Ligands: CIT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-12-11 Classification: HYDROLASE Ligands: SO4 |
 |
Step (Ptpn5) With Active-Site Disulfide Bond And Covalent Ligand Bound To Distal C505 And C518
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-12-11 Classification: HYDROLASE Ligands: A1BHT, DMS, SO4 |
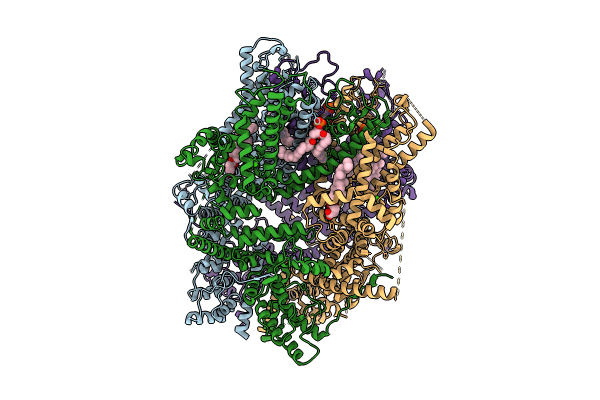 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: METAL TRANSPORT Ligands: LPP, ZN, Y01, CA |
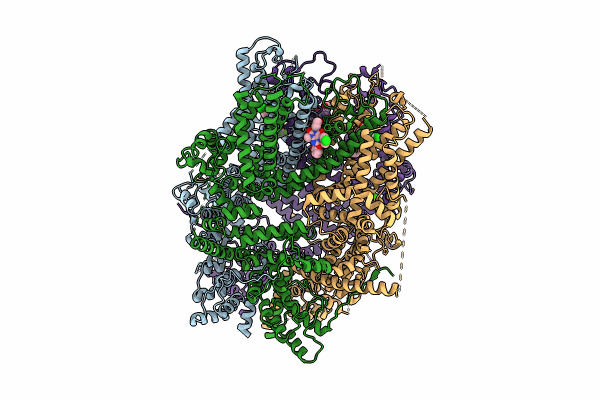 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: METAL TRANSPORT Ligands: PJQ, ZN, CA |
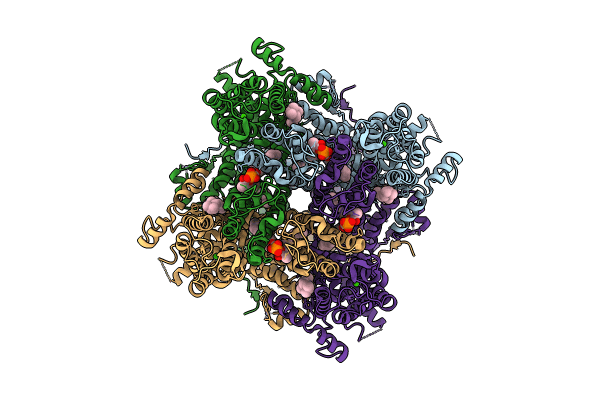 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: METAL TRANSPORT Ligands: Y01, ZN, CA, LPP |
 |
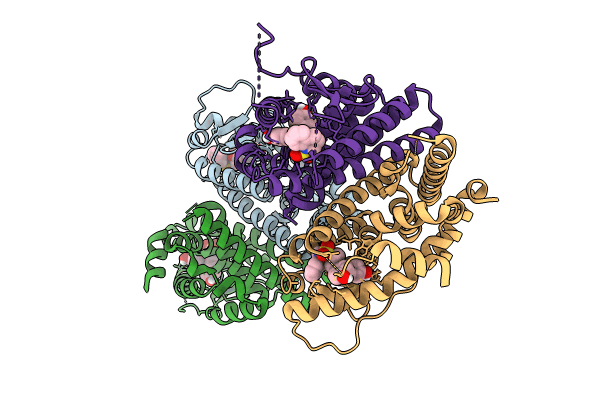
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-411
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHV |
 |
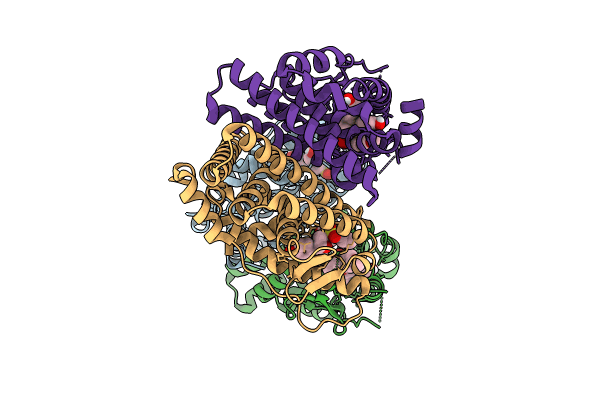
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-410
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHU |
 |
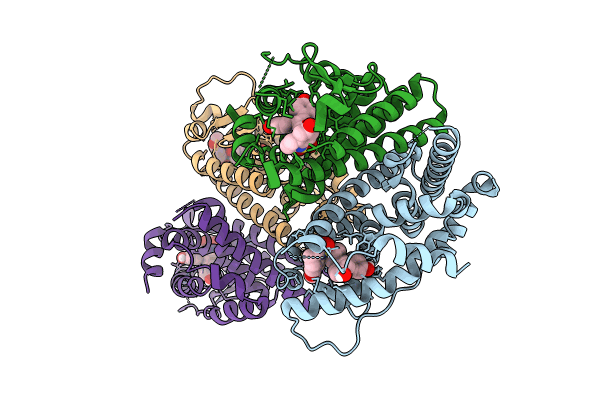
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-400
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHO |
 |
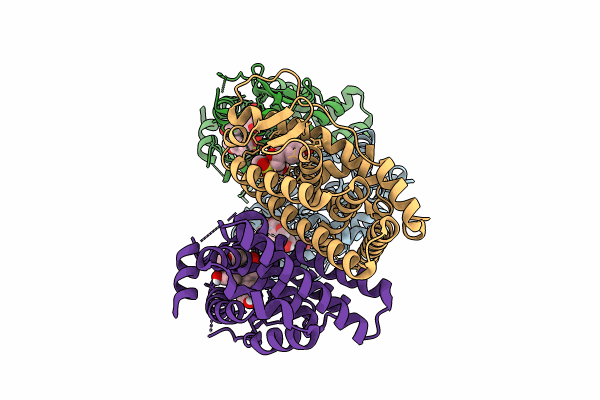
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-409
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHS |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-403
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHW |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-406
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHX, NI |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-1154
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: OBT |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-402
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHY, A1AHZ |
 |
Pseudouridine 5'-Monophosphate Glycosylase From Arabidopsis Thaliana -- Sulfate Bound Holoenzyme
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2024-05-15 Classification: HYDROLASE Ligands: MN, SO4 |
 |
Pseudouridine 5'-Monophosphate Glycosylase From Arabidopsis Thaliana -- Psu, R5P Bound K185A Mutant
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-05-15 Classification: HYDROLASE Ligands: MN, PSU, RP5 |
 |
Pseudouridine 5'-Monophosphate Glycosylase From Arabidopsis Thaliana -- Citrate Bound K185A Mutant
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.12 Å Release Date: 2024-05-15 Classification: HYDROLASE Ligands: MN, CIT |
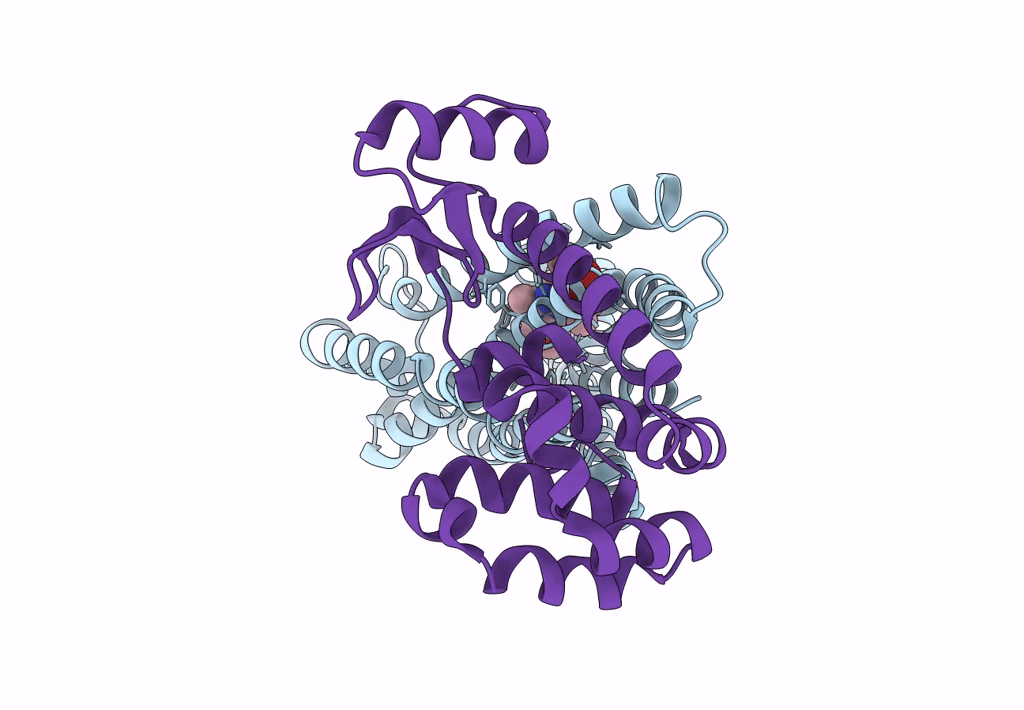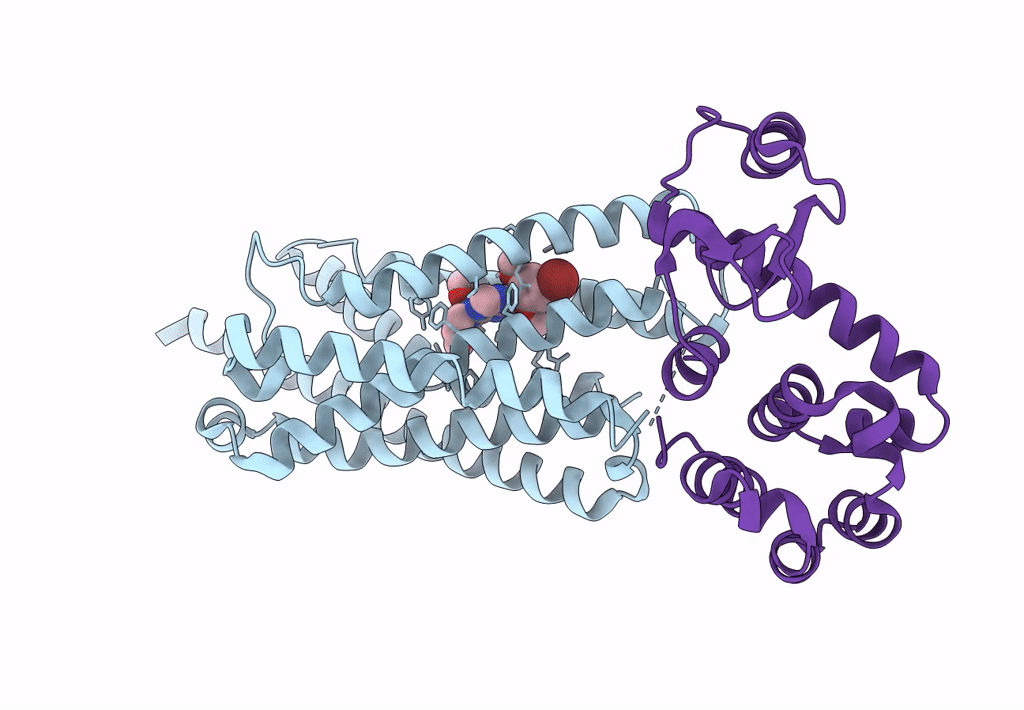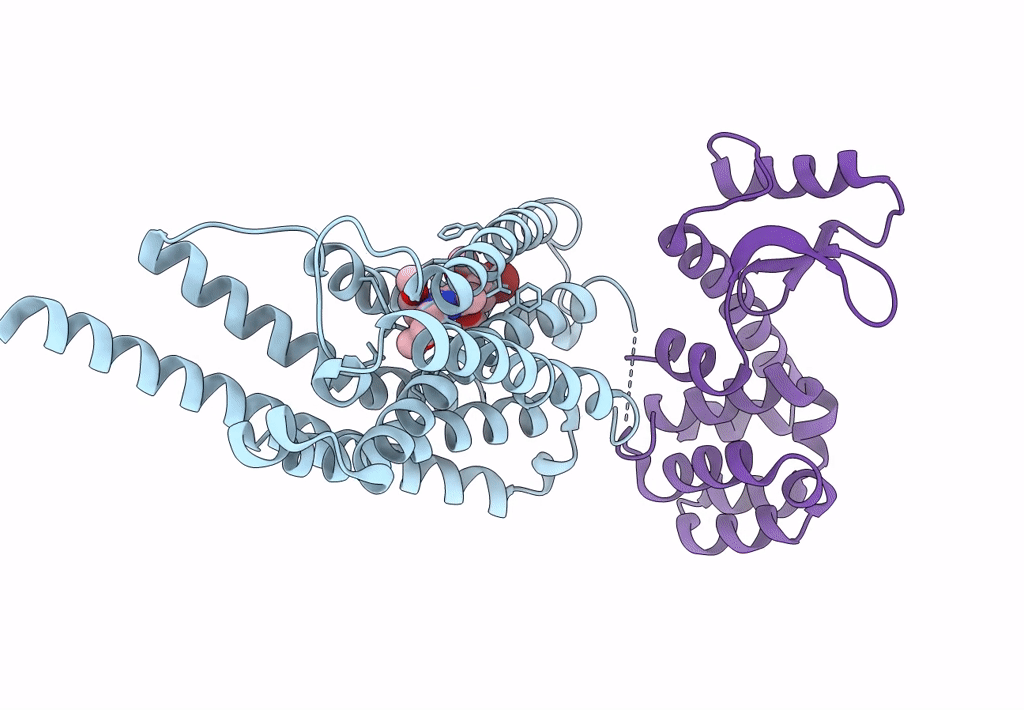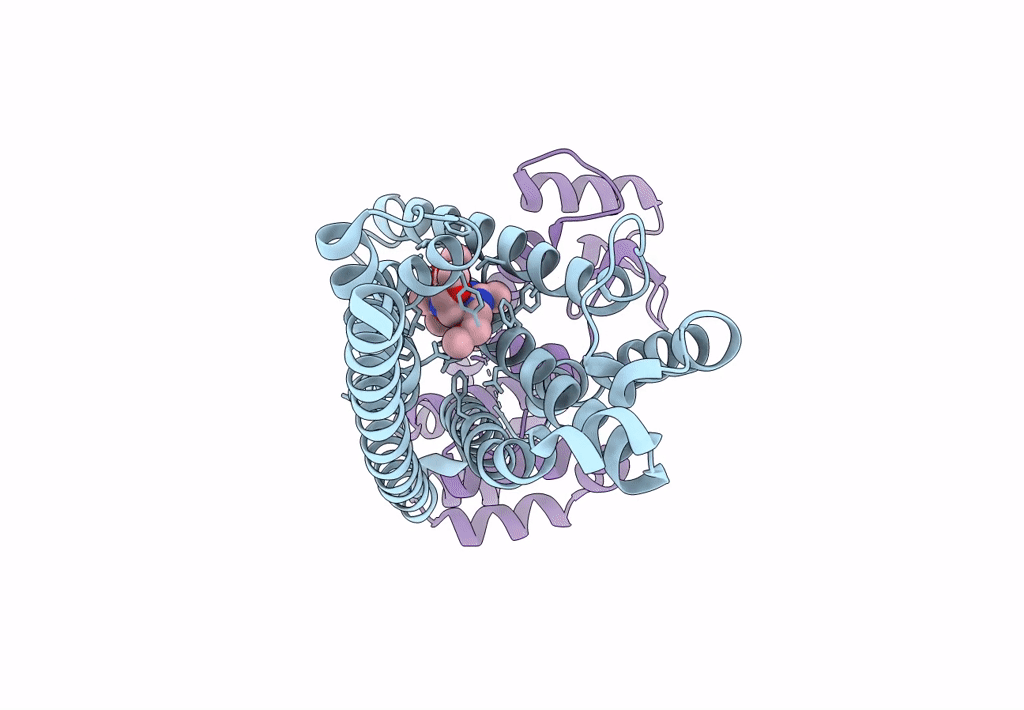 |
Organism: Homo sapiens, Enterobacteria phage t6
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2023-09-13 Classification: MEMBRANE PROTEIN Ligands: CW9 |
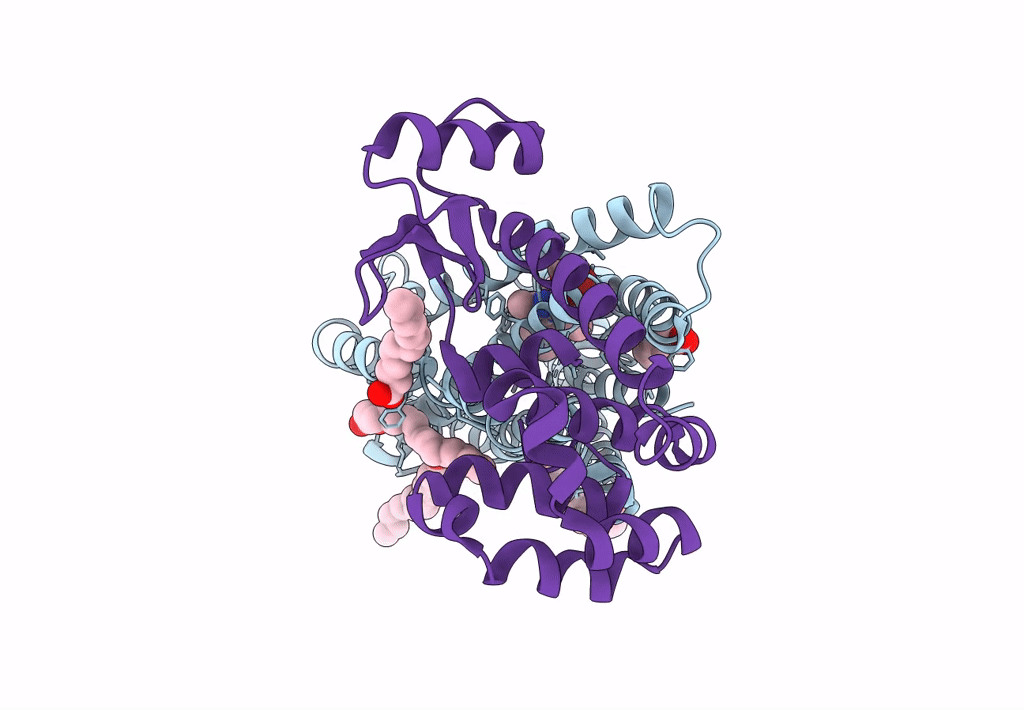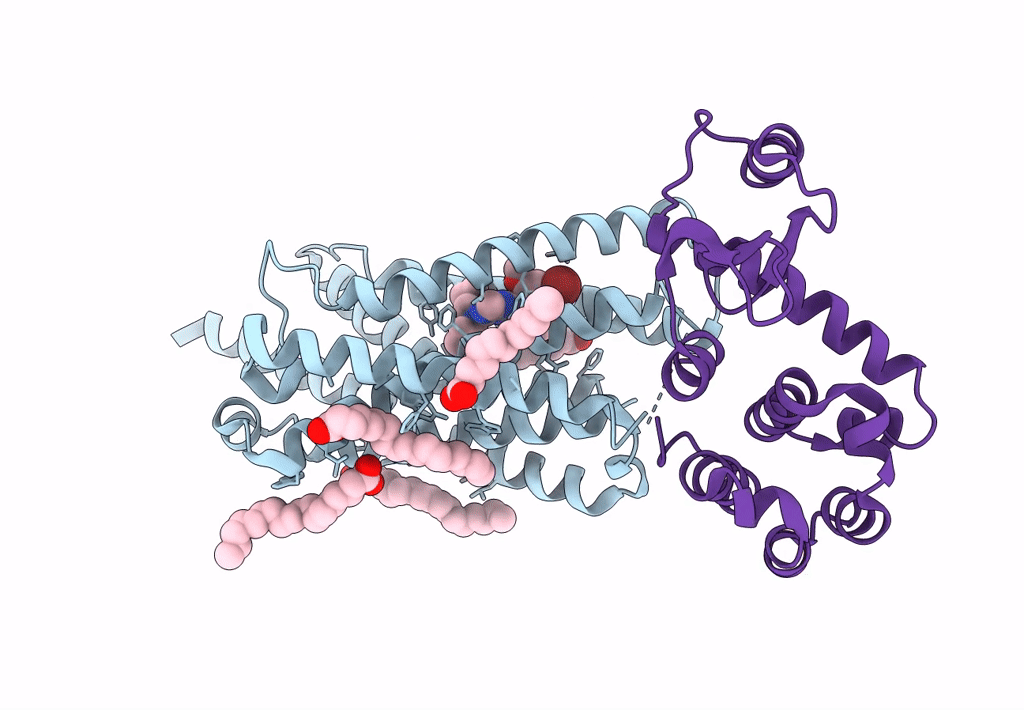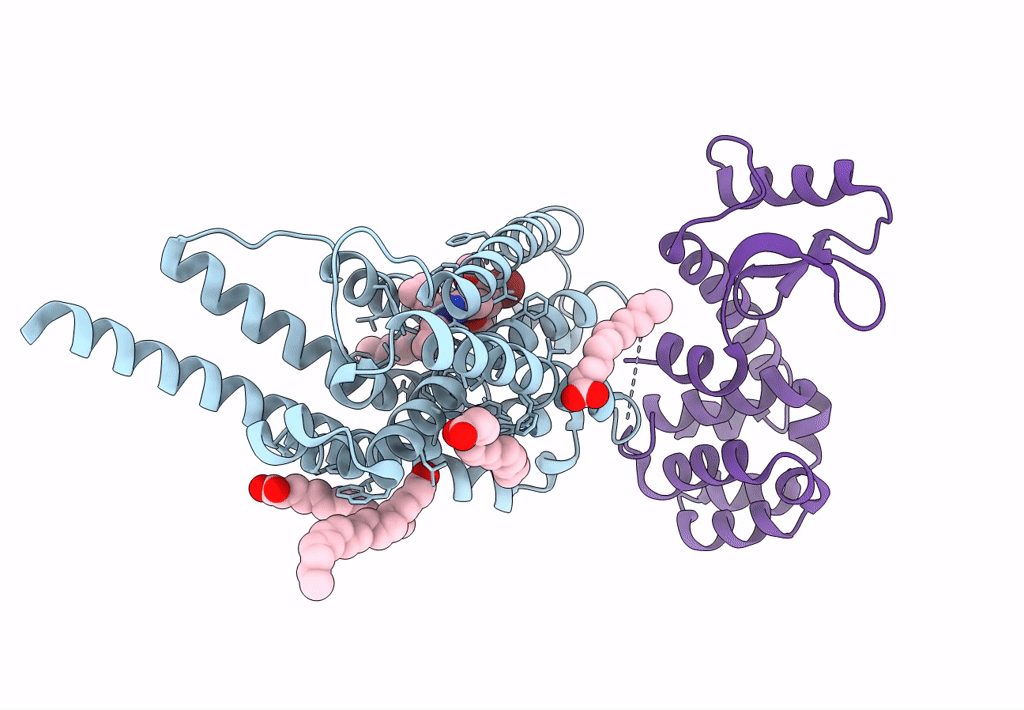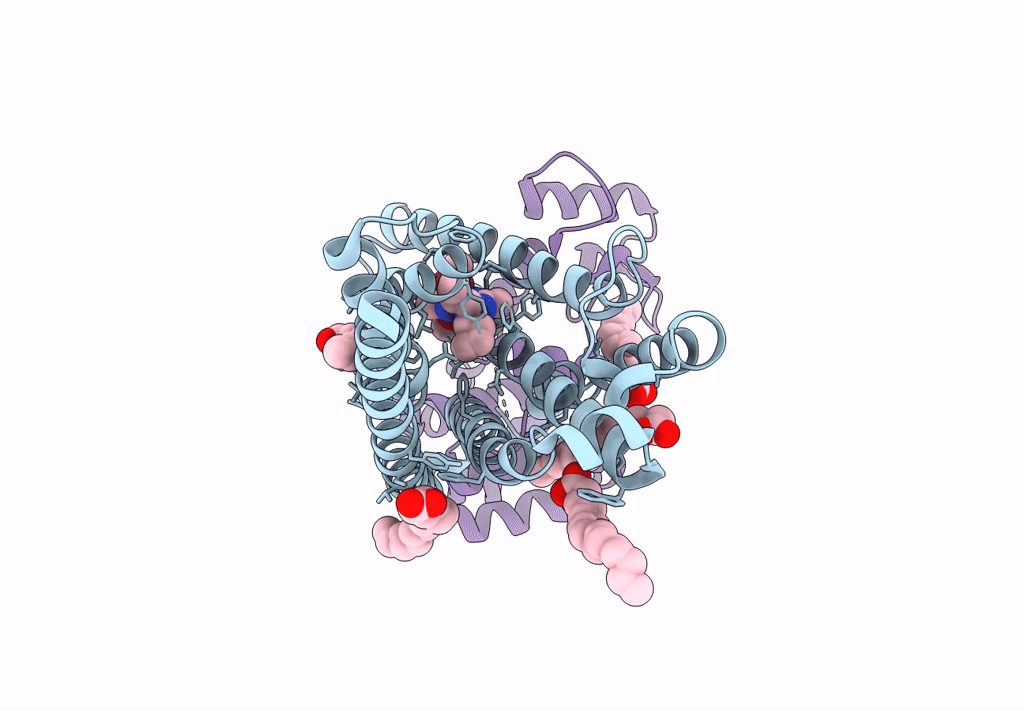 |
Organism: Homo sapiens, Enterobacteria phage t6
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-09-13 Classification: MEMBRANE PROTEIN Ligands: OLA, 0JS |
 |
Organism: Homo sapiens, Enterobacteria phage t6
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-09-13 Classification: MEMBRANE PROTEIN Ligands: 0VI |

