Search Count: 28
 |
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2022-06-01 Classification: IMMUNE SYSTEM |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-12-02 Classification: ANTIBIOTIC Ligands: UQ8 |
 |
Organism: Listeria monocytogenes
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-06-05 Classification: DNA BINDING PROTEIN Ligands: CL |
 |
Organism: Listeria monocytogenes
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-06-05 Classification: DNA BINDING PROTEIN |
 |
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-05-01 Classification: DNA BINDING PROTEIN |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-06-27 Classification: CHAPERONE |
 |
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-03-08 Classification: TRANSCRIPTION REGULATOR Ligands: PO4, GLC, IPA |
 |
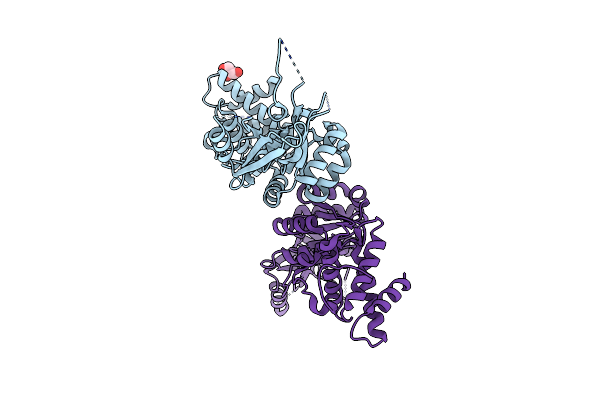
Organism: Pseudomonas aeruginosa (strain atcc 15692 / pao1 / 1c / prs 101 / lmg 12228)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-08-10 Classification: TRANSFERASE Ligands: TRS, MG, PO4 |
 |
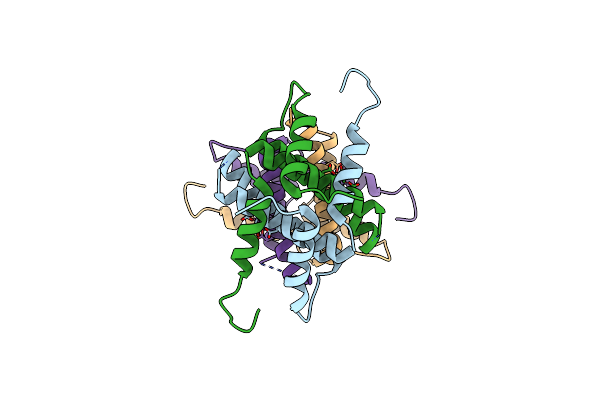
Organism: Thermococcus onnurineus (strain na1)
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2016-05-11 Classification: HYDROLASE Ligands: GOL |
 |
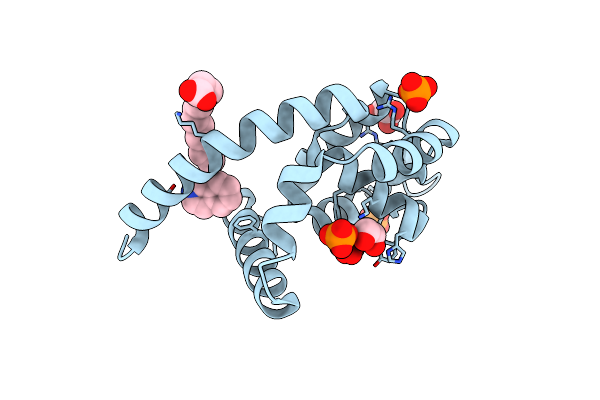
Crystal Structure Of The Bacillus-Conserved Mazg Protein, A Nucleotide Pyrophosphohydrolase
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-03-30 Classification: HYDROLASE Ligands: MN |
 |
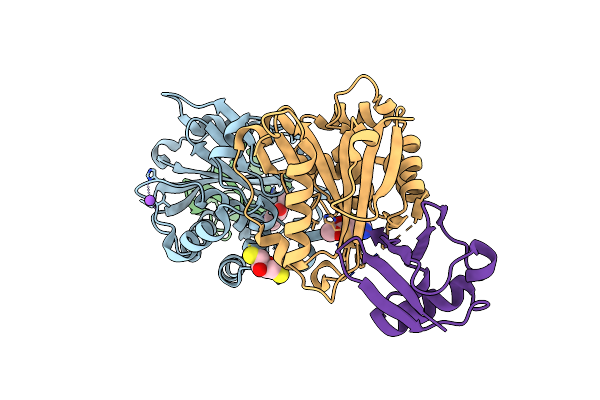
Organism: Bacillus cereus (strain atcc 14579 / dsm 31)
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2016-02-10 Classification: TRANSCRIPTION Ligands: PO4, ISQ, DAO, GOL, NI, SO4 |
 |
Crystal Structure Of Trichinella Spiralis Uch37 Catalytic Domain Bound To Ubiquitin Vinyl Methyl Ester
Organism: Trichinella spiralis, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-05-29 Classification: Hydrolase/Signaling Protein Ligands: DTT, NA, GVE |
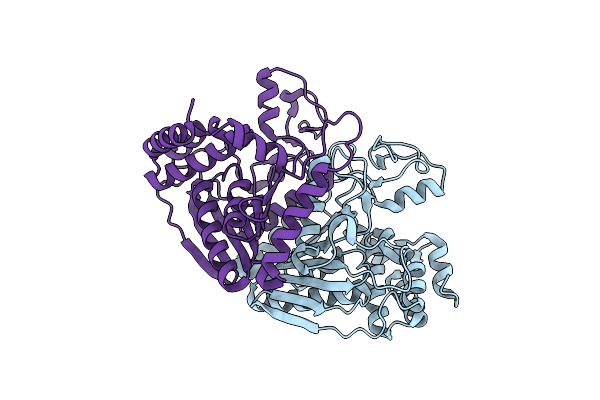 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2013-01-16 Classification: OXIDOREDUCTASE |
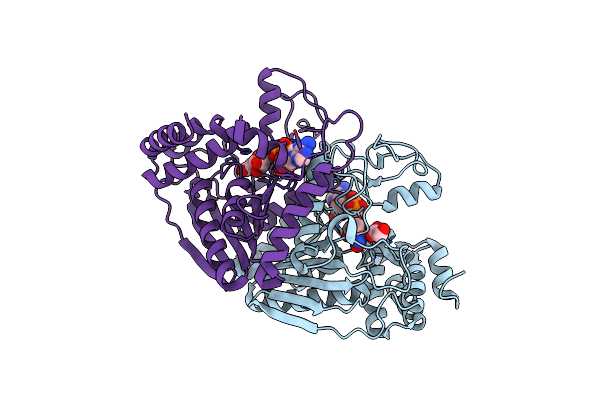 |
Crystal Structure Of Ureidoglycolate Dehydrogenase In Ternary Complex With Nadh And Glyoxylate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2013-01-16 Classification: OXIDOREDUCTASE Ligands: NAI, GLV |
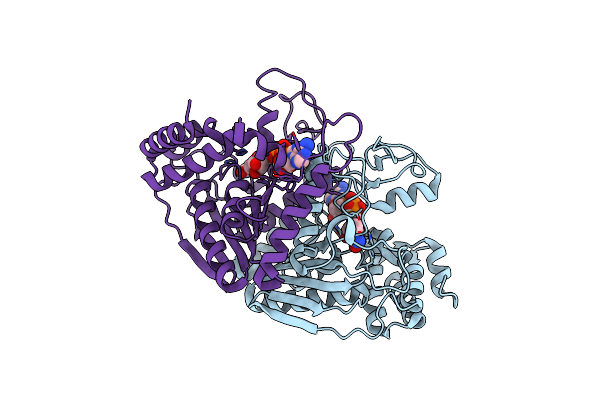 |
Crystal Structure Of Ureidoglycolate Dehydrogenase In Binary Complex With Nadh
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2013-01-16 Classification: OXIDOREDUCTASE Ligands: NAI |
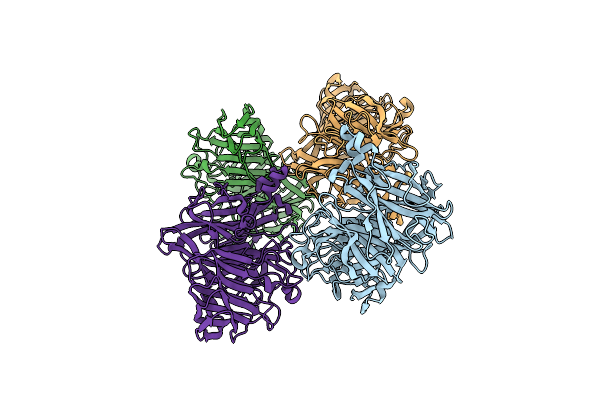 |
Organism: Arthrobacter ureafaciens
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2012-07-18 Classification: HYDROLASE |
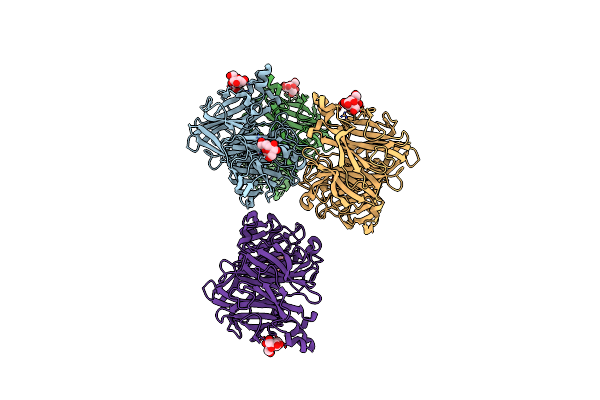 |
Crystal Structure Of Levan Fructotransferase From Arthrobacter Ureafaciens In Complex With Dfa-Iv
Organism: Arthrobacter ureafaciens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-07-18 Classification: TRANSFERASE Ligands: 0U8 |
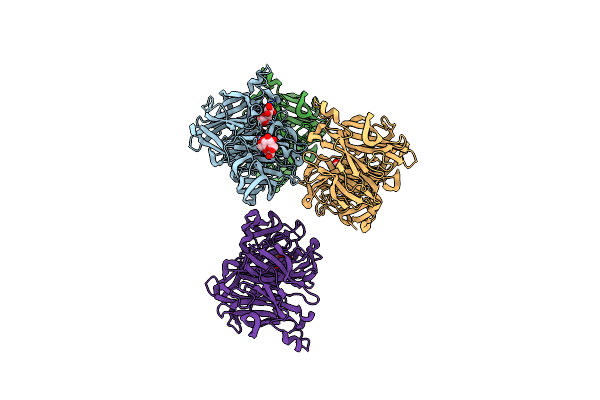 |
Crystal Structure Of Levan Fructotransferase D54N Mutant From Arthrobacter Ureafaciens In Complex With Sucrose
Organism: Arthrobacter ureafaciens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-07-18 Classification: TRANSFERASE |
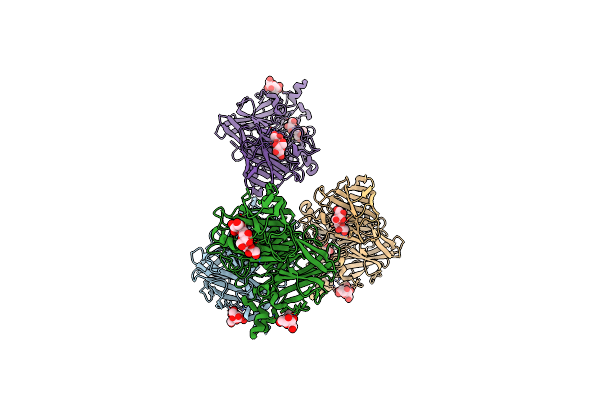 |
Crystal Structure Of Levan Fructotransferase D54N Mutant From Arthrobacter Ureafaciens In Complex With Levanbiose
Organism: Arthrobacter ureafaciens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-07-18 Classification: TRANSFERASE |
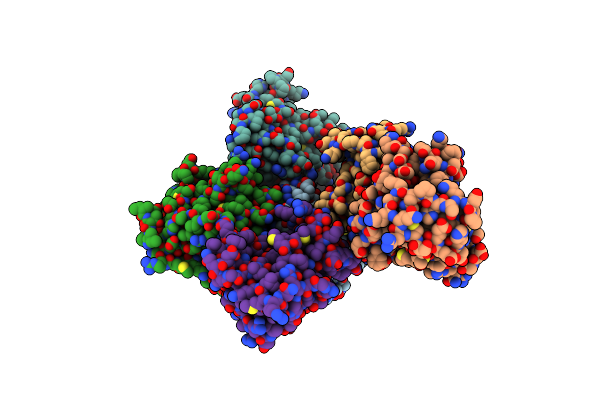 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-10-19 Classification: HYDROLASE Ligands: ZN |

