Search Count: 67
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2024-09-25 Classification: STRUCTURAL PROTEIN |
 |
Crystal Structure Of Vibrio Vulnificus Rid-Dependent Transforming Nadase Domain (Rdtnd)/Calmodulin-Binding Domain Of Rho Inactivation Domain (Rid-Cbd) Complexed With Ca2+-Bound Calmodulin
Organism: Vibrio vulnificus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-07-10 Classification: TOXIN Ligands: CA |
 |
Crystal Structure Of Vibrio Vulnificus Rid-Dependent Transforming Nadase Domain (Rdtnd)/Calmodulin-Binding Domain Of Rho Inactivation Domain (Rid-Cbd) Complexed With Ca2+-Bound Calmodulin And A Nicotinamide Adenine Dinucleotide (Nad+)
Organism: Vibrio vulnificus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2024-07-10 Classification: TOXIN Ligands: NAD, CA, GOL |
 |
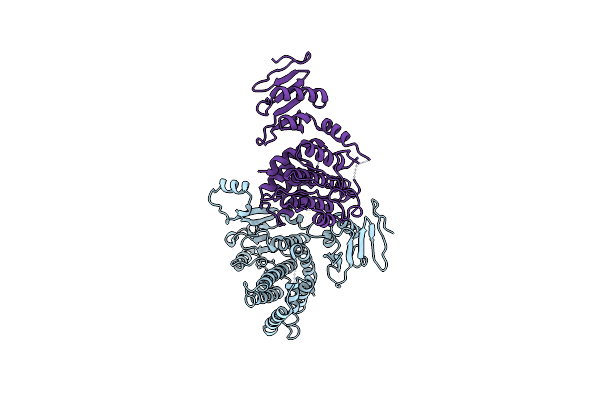
Crystal Structure Of Vibrio Vulnificus Rid-Dependent Transforming Nadase Domain (Rdtnd)/Calmodulin-Binding Domain Of Rho Inactivation Domain (Rid-Cbd) Complexed With Ca2+-Free Calmodulin
Organism: Vibrio vulnificus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2024-07-10 Classification: TOXIN Ligands: MG |
 |
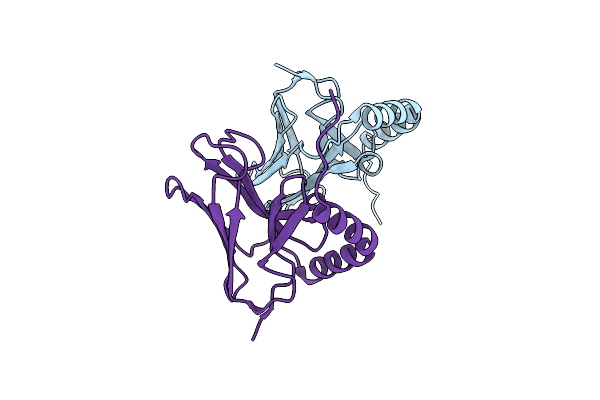
Crystal Structure Of The Rid-Dependent Transforming Nadase Domain (Rdtnd)/Calmodulin-Binding Domain Of Rho Inactivation Domain (Rid-Cbd) From Vibrio Vulnificus
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:3.38 Å Release Date: 2024-07-10 Classification: TOXIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2019-10-02 Classification: CHAPERONE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-10-02 Classification: CHAPERONE |
 |
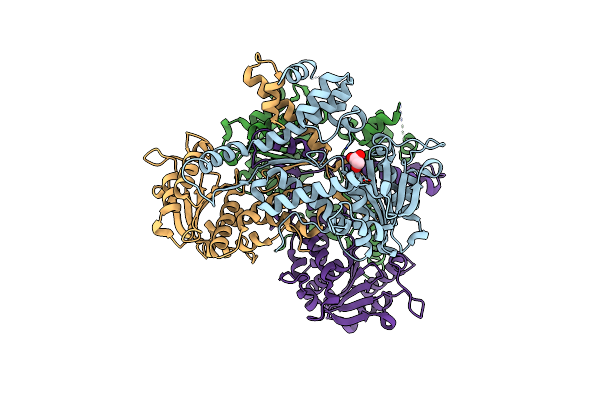
Organism: Vibrio vulnificus mo6-24/o
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2019-08-21 Classification: TOXIN |
 |
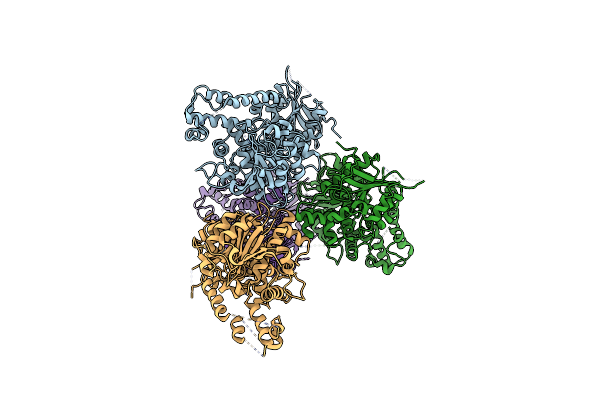
Crystal Structure Of The Makes Caterpillars Floppy (Mcf)-Like Effector Of Vibrio Vulnificus Mo6-24/O
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2019-08-07 Classification: TOXIN Ligands: GOL |
 |
Crystal Structure Of Alpha-Beta Hydrolase (Abh) And Makes Caterpillars Floppy (Mcf)-Like Effectors Of Vibrio Vulnificus Mo6-24/O
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2019-08-07 Classification: TOXIN |
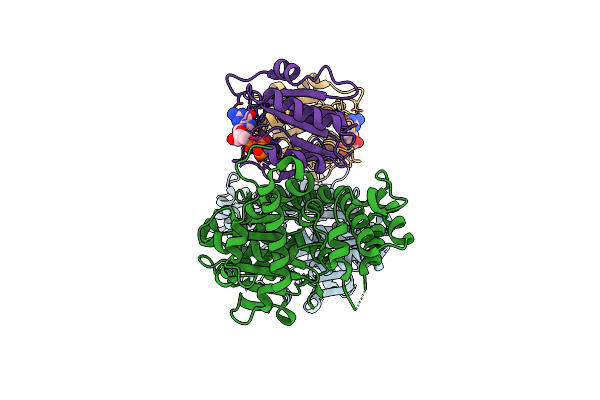 |
Crystal Structure Of The Makes Caterpillars Floppy (Mcf)-Like Effector Of Vibrio Vulnificus Mo6-24/O In Complex With A Human Adp-Ribosylation Factor 3 (Arf3)
Organism: Vibrio vulnificus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-08-07 Classification: TOXIN/PROTEIN BINDING Ligands: GTP, MG |
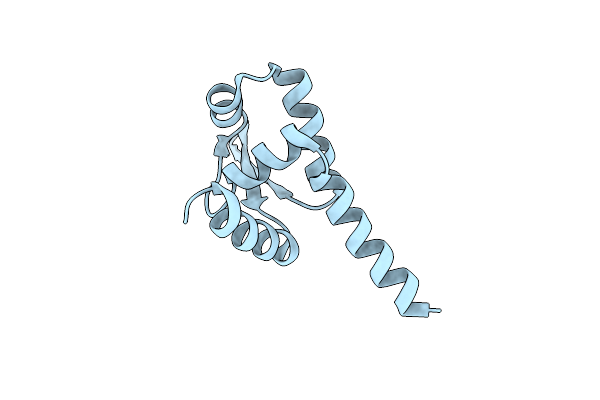 |
Organism: Vibrio vulnificus cmcp6
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2019-04-10 Classification: DNA BINDING PROTEIN |
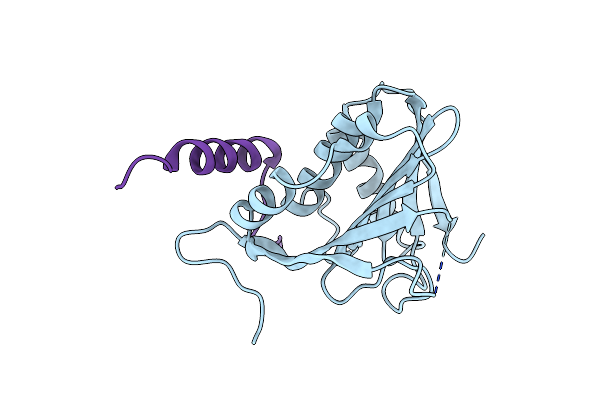 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-11-21 Classification: RNA BINDING PROTEIN/LIGASE |
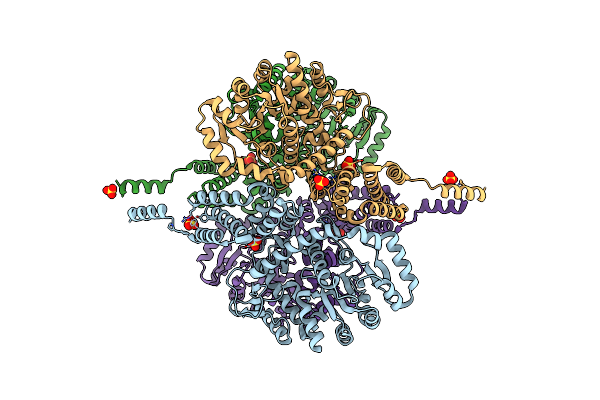 |
Organism: Vibrio vulnificus cmcp6
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-10-10 Classification: TOXIN Ligands: SO4 |
 |
Organism: Vibrio vulnificus cmcp6
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2018-10-10 Classification: TOXIN Ligands: SO4, GOL |
 |
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-01-24 Classification: DNA BINDING PROTEIN Ligands: 7Y3, SO4 |
 |
Organism: Pseudomonas putida (strain f1 / atcc 700007)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-03-02 Classification: TRANSFERASE |
 |
Organism: Pseudomonas putida (strain f1 / atcc 700007)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2016-03-02 Classification: TRANSFERASE Ligands: MBN |
 |
Organism: Pseudomonas putida (strain f1 / atcc 700007)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-03-02 Classification: TRANSFERASE Ligands: XBZ |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2015-12-23 Classification: OXIDOREDUCTASE Ligands: BEZ, CL, GOL |

