Search Count: 550
 |
The L2A-I Alpha-Synuclein Fibril In The Presence Of Modag-005 (Short Incubation)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: PROTEIN FIBRIL |
 |
The L2A-Ii Alpha-Synuclein Fibril In The Presence Of Modag-005 (Short Incubation)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: PROTEIN FIBRIL |
 |
The L2C Alpha-Synuclein Fibril In The Presence Of Modag-005 (Short Incubation)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: PROTEIN FIBRIL |
 |
The L2A-I Alpha-Synuclein Fibril In The Presence Of Modag-005 (Long Incubation)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: PROTEIN FIBRIL |
 |
The L2A-Ii Alpha-Synuclein Fibril In The Presence Of Modag-005 (Long Incubation)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: PROTEIN FIBRIL |
 |
The L2C Alpha-Synuclein Fibril In The Presence Of Modag-005 (Long Incubation)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: PROTEIN FIBRIL |
 |
Organism: Bacillus subtilis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: DNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: CHAPERONE Ligands: ATP, K |
 |
Organism: Ralstonia solanacearum uw551
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: IMMUNOSUPPRESSANT |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: MEMBRANE PROTEIN |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens, Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: CYTOSOLIC PROTEIN/CONTRACTILE PROTEIN |
 |
Organism: Homo sapiens, Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: CYTOSOLIC PROTEIN/CONTRACTILE PROTEIN |
 |
Cryo-Em Structure Of Outward State Anhydromuropeptide Permease (Ampg) Complex With Glcnac-1,6-Anhmurnac
Organism: Homo sapiens, Synthetic construct, Yokenella regensburgei, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: MEMBRANE PROTEIN Ligands: 2YP |
 |
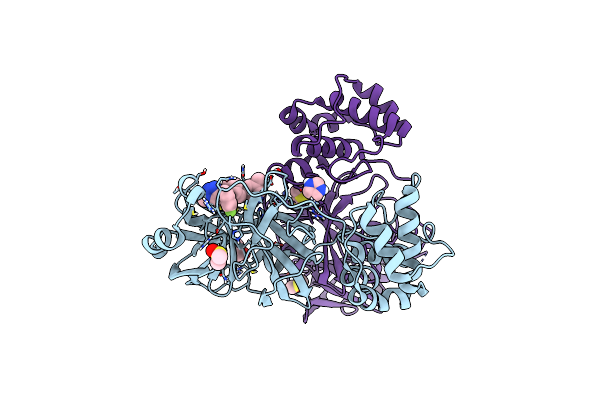
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro) In Complex With The Noncovalently Bound Inhibitor C5N17A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: VIRAL PROTEIN Ligands: DMS, A1IHT, CL, MG |
 |
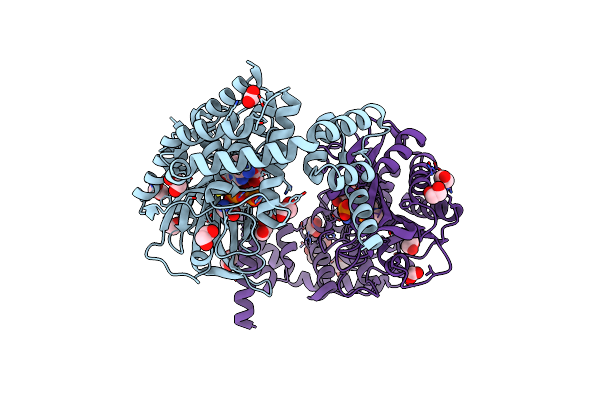
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro) In Complex With The Noncovalently Bound Inhibitor C5N17B
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: VIRAL PROTEIN Ligands: DMS, A1IHV, IMD |
 |
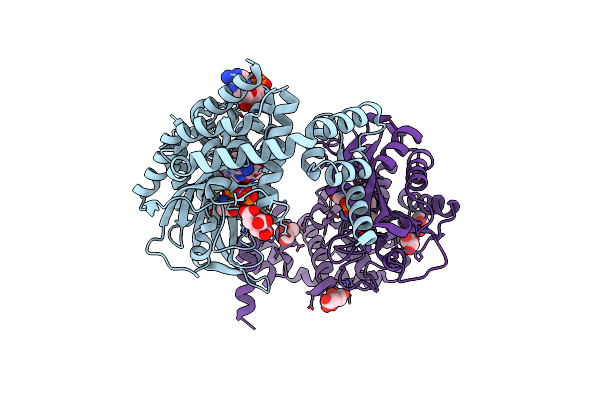
Crystal Structure Of Alanyl-Trna Synthetase In Complex With Atp And L-Alanine
Organism: Methylomonas sp. dh-1
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: LIGASE Ligands: ATP, ALA, EDO, FMT, MLT, CIT |
 |
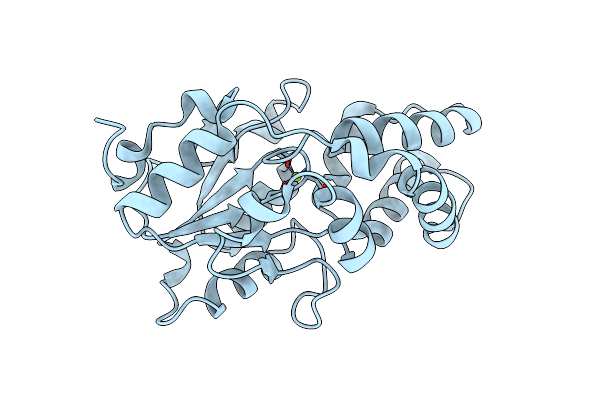
Crystal Structure Of Alanyl-Trna Synthetase L219M Mutant In Complex With Atp And L-Alanine
Organism: Methylomonas sp. dh-1
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: LIGASE Ligands: ATP, ALA, CIT, TRS, GOL |
 |
Crystal Structure Of Pseudouridine 5'-Monophosphate Phosphatase From Human (Hhdhd1A) In The Unliganded State
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: RNA BINDING PROTEIN Ligands: MG |
 |
Crystal Structure Of Human Pseudouridine 5'-Monophosphate Phosphatase (Hhdhd1A) Complexed With Pseudouridine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: RNA BINDING PROTEIN Ligands: MG, FJF |

