Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Micromonospora pattaloongensis
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2025-01-15 Classification: HYDROLASE Ligands: EDO, GOL |
 |
Organism: Micromonospora pattaloongensis
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2025-01-15 Classification: HYDROLASE |
 |
Organism: Kutzneria buriramensis
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2025-01-15 Classification: HYDROLASE |
 |
Organism: Kutzneria buriramensis
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2025-01-15 Classification: HYDROLASE |
 |
Organism: Kutzneria buriramensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-01-15 Classification: HYDROLASE Ligands: ZN, 1PE, ACT |
 |
Organism: Aspergillus fumigatiaffinis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-05-15 Classification: HYDROLASE |
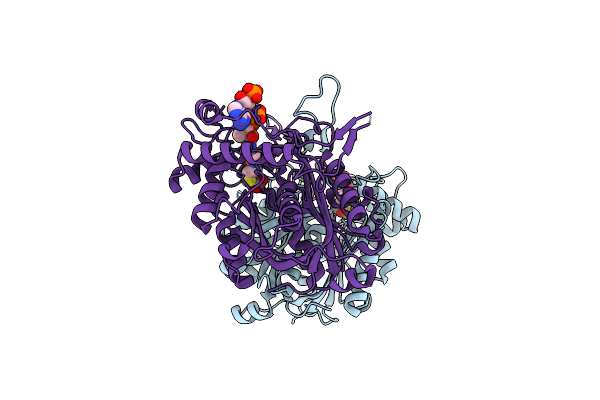 |
Crystal Structure Of A Biosynthetic Thiolase From Megasphaera Hexanoica Soaked With Hexanoyl-Coa
Organism: Megasphaera hexanoica
Method: X-RAY DIFFRACTION Release Date: 2024-05-08 Classification: BIOSYNTHETIC PROTEIN Ligands: COA, ACT |
 |
Organism: Clostridium kluyveri
Method: X-RAY DIFFRACTION Release Date: 2024-05-08 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-03-13 Classification: TRANSFERASE Ligands: GOL |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-03-13 Classification: TRANSFERASE |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-03-13 Classification: TRANSFERASE Ligands: GOL |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2024-03-13 Classification: TRANSFERASE Ligands: GOL |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2024-03-13 Classification: TRANSFERASE Ligands: GOL, PGE, 2PE |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2024-03-13 Classification: TRANSFERASE Ligands: CIT |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-03-13 Classification: TRANSFERASE Ligands: GOL |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2024-03-13 Classification: TRANSFERASE Ligands: ACO, GOL |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-03-13 Classification: TRANSFERASE Ligands: COA |
 |
Organism: Lederbergvirus
Method: ELECTRON MICROSCOPY Release Date: 2024-01-31 Classification: VIRUS LIKE PARTICLE |
 |
Crystal Structure Of Acetylornithine Aminotransferase From Corynebacterium Glutamicum
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-01-17 Classification: TRANSFERASE |
 |
Crystal Structure Of Acetylornithine Aminotransferase Complex With Plp From Corynebacterium Glutamicum
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2024-01-17 Classification: TRANSFERASE Ligands: PLP |