Search Count: 87
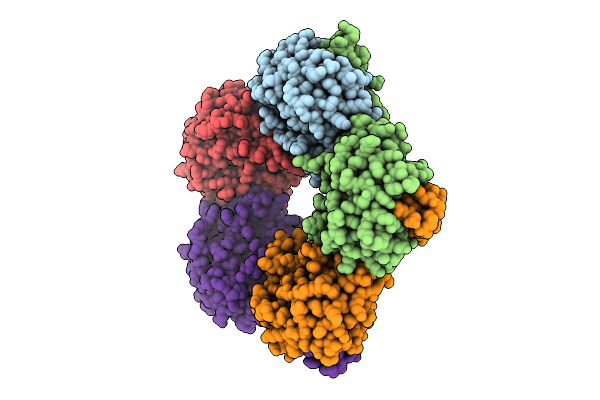 |
Selective Production Of Versatile L-Glyceraldehyde From C1 And/Or C2 Aldehydes
Organism: Aeromonas sp. asnih1
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: SUGAR BINDING PROTEIN |
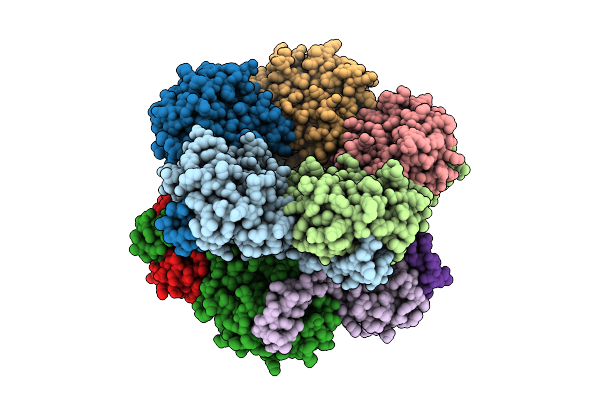 |
Selective Production Of Versatile L-Glyceraldehyde From C1 And/Or C2 Aldehydes
Organism: Gilliamella apicola
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: SUGAR BINDING PROTEIN |
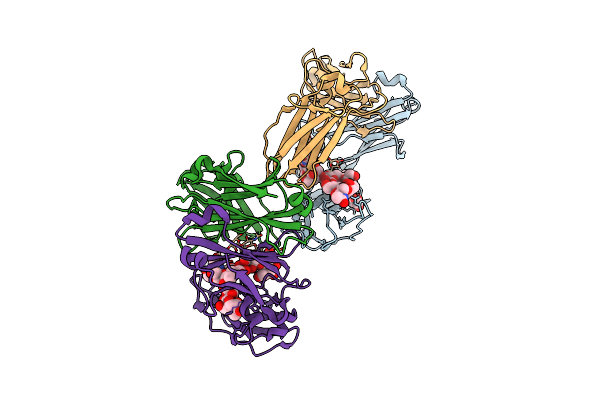 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: IMMUNE SYSTEM Ligands: GAL |
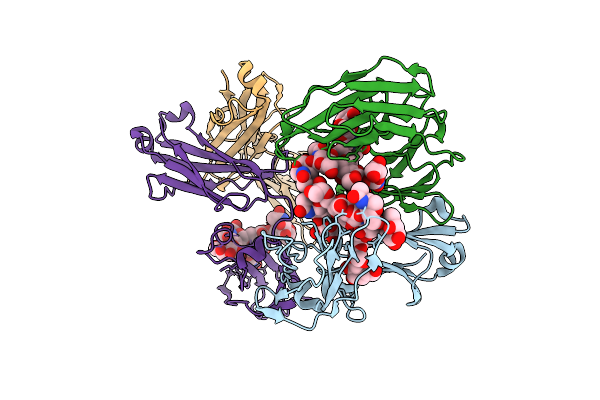 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: IMMUNE SYSTEM Ligands: FUL |
 |
Cryo-Em Structure Of Outward State Anhydromuropeptide Permease (Ampg) Complex With Glcnac-1,6-Anhmurnac
Organism: Homo sapiens, Synthetic construct, Yokenella regensburgei, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: MEMBRANE PROTEIN Ligands: 2YP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: IMMUNE SYSTEM |
 |
Organism: Yokenella regensburgei
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: TRANSPORT PROTEIN |
 |
Cryo-Em Structure Of Inward-Facing Anhydromuropeptide Permease (Ampg) In Complex With Glcnac-1,6-Anhmurnac
Organism: Yokenella regensburgei
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: MEMBRANE PROTEIN Ligands: 2YP |
 |
Cryo-Em Structure Of Outward State Anhydromuropeptide Permease (Ampg) G50W/L269W
Organism: Yokenella regensburgei, Escherichia coli, Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: PEPTIDE BINDING PROTEIN |
 |
Organism: Klebsiella pneumoniae, Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: STRUCTURAL PROTEIN/IMMUNE SYSTEM Ligands: CIT, PLM, NAG |
 |
Organism: Klebsiella pneumoniae, Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: STRUCTURAL PROTEIN/IMMUNE SYSTEM Ligands: CIT, PLM, NAG |
 |
Cryo-Em Structure Of The G15C-R66C And T83C-T83C Diabody Complex (Cits-Diabody #7-Tlr3)
Organism: Klebsiella pneumoniae, Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: STRUCTURAL PROTEIN/IMMUNE SYSTEM Ligands: CIT, PLM, NAG |
 |
Cryo-Em Structure Of The Klebsiella Pneumoniae Cits (Citrate-Bound Occluded State)
Organism: Klebsiella pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: MEMBRANE PROTEIN Ligands: CIT, PLM |
 |
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: MEMBRANE PROTEIN Ligands: PTY, NAG |
 |
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: MEMBRANE PROTEIN Ligands: PTY, NAG |
 |
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: MEMBRANE PROTEIN Ligands: PTY |
 |
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: MEMBRANE PROTEIN Ligands: 4DS, PTY, NAG |
 |
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: MEMBRANE PROTEIN Ligands: PTY, NAG, 4DS |

