Search Count: 136
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Pfr State (I0A).
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL, EDO |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Pfr State (I0B).
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4 |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I1 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4 |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I2 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, PGE, PEG, CL |
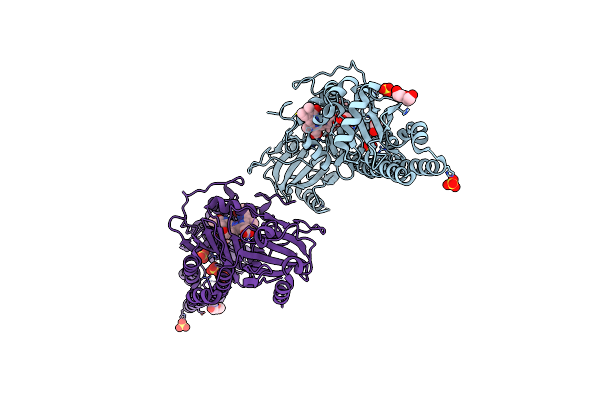 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I3 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, GOL, PEG |
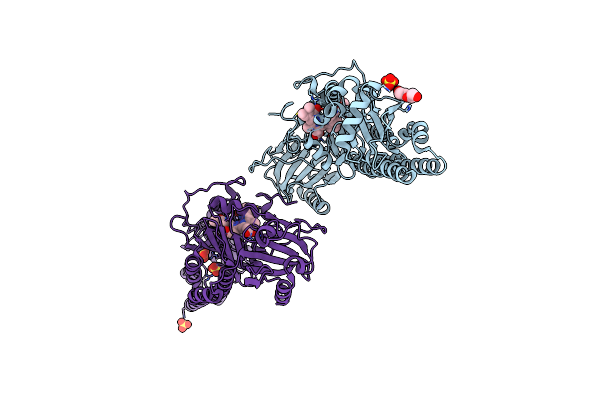 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I4 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL, PEG |
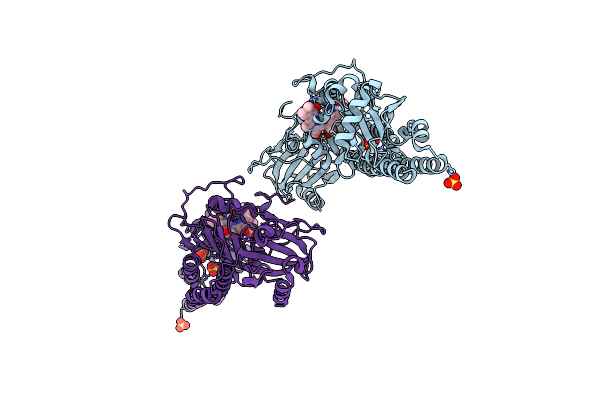 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I5 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL |
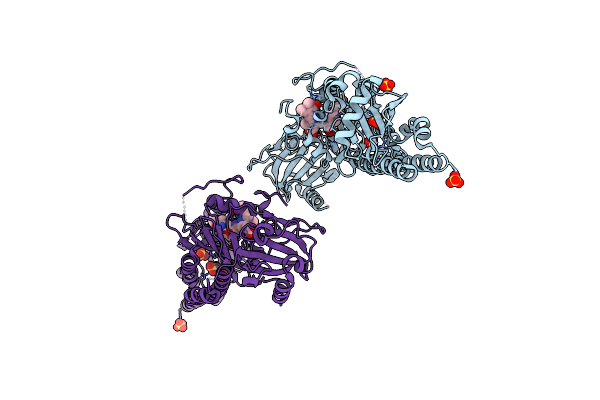 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I6 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I7 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, PEG |
 |
E. Coli Dhfr Complex With Nadp+ And Folate: Ef-X Off Model By Laue Diffraction (No Electric Field)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-01-03 Classification: OXIDOREDUCTASE Ligands: FOL, NAP, MN |
 |
E. Coli Dhfr Complex With Nadp+ And Folate: Ef-X Excited State Model By Laue Diffraction (Electric Field Along B Axis; 8-Fold Extrapolation Of Structure Factor Differences)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-01-03 Classification: OXIDOREDUCTASE Ligands: FOL, NAP, MN |
 |
Time-Resolved Sfx-Xfel Crystal Structure Of Cyp121 Bound With Cyy Reacted With Peracetic Acid For 200 Milliseconds
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: SO4, HEM, YTT, PEO |
 |
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: SO4, HEM, YTT |
 |
Ribonucleotide Reductase Class Ie R2 From Mesoplasma Florum, Catalytically Active Radical State Solved By Xfel
Organism: Mesoplasma florum l1
Method: X-RAY DIFFRACTION Release Date: 2023-11-01 Classification: OXIDOREDUCTASE |
 |
Ribonucleotide Reductase Class Ie R2 From Mesoplasma Florum, Radical-Lost Ground State
Organism: Mesoplasma florum l1
Method: X-RAY DIFFRACTION Release Date: 2023-11-01 Classification: OXIDOREDUCTASE Ligands: CA, GOL |
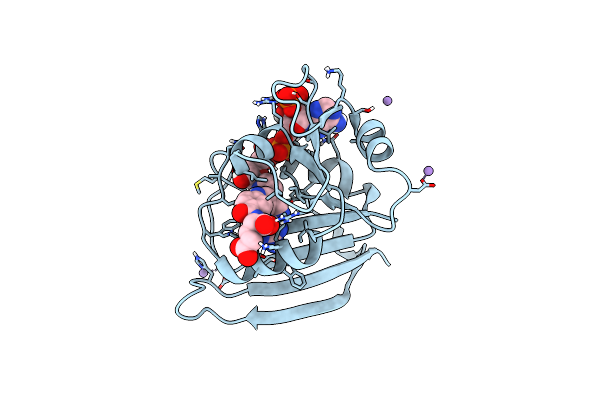 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2023-09-20 Classification: OXIDOREDUCTASE Ligands: FOL, NAP, MN |
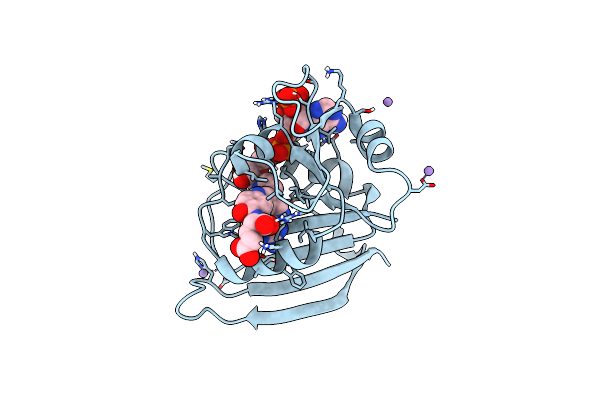 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.04 Å Release Date: 2023-09-20 Classification: OXIDOREDUCTASE Ligands: FOL, NAP, MN |
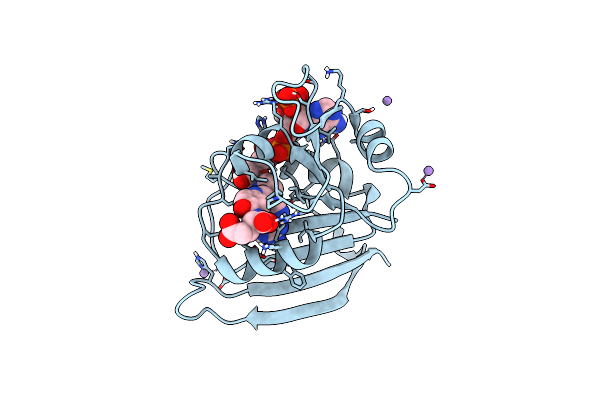 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.04 Å Release Date: 2023-09-20 Classification: OXIDOREDUCTASE Ligands: FOL, NAP, MN |
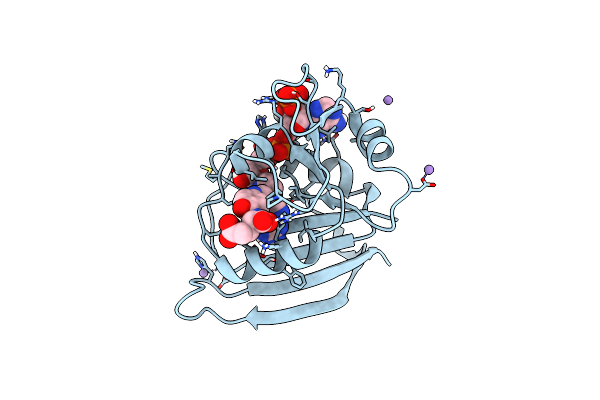 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2023-09-20 Classification: OXIDOREDUCTASE Ligands: FOL, NAP, MN |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.03 Å Release Date: 2023-09-20 Classification: OXIDOREDUCTASE Ligands: FOL, NAP, MN |

