Search Count: 20
 |
Organism: Thermoascus crustaceus
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: HYDROLASE |
 |
Organism: Cellulomonas biazotea
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-05-28 Classification: HYDROLASE Ligands: GOL |
 |
Crystal Structure Of Beta-Glucosidase Cba3 From Cellulomonas Biazotea In Complex With Glucose
Organism: Cellulomonas biazotea
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-05-28 Classification: HYDROLASE Ligands: BGC |
 |
Organism: Montipora sp. 20
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-04-23 Classification: FLUORESCENT PROTEIN |
 |
Organism: Thermoanaerobacterium saccharolyticum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-11-22 Classification: HYDROLASE Ligands: ACT |
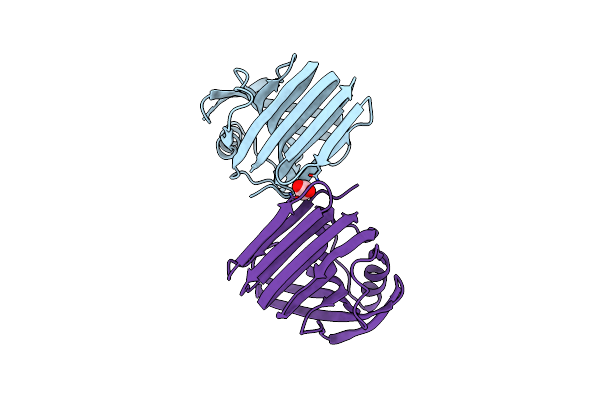 |
Room Temperature Structure Of Gh11 From Thermoanaerobacterium Saccharolyticum By Serial Crystallography
Organism: Thermoanaerobacterium saccharolyticum
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2023-11-22 Classification: HYDROLASE Ligands: ACT |
 |
Crystal Structure Of Beta-Glucosidase From Thermoanaerobacterium Saccharolyticum
Organism: Thermoanaerobacterium saccharolyticum (strain dsm 8691 / jw/sl-ys485)
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2022-02-23 Classification: HYDROLASE Ligands: TRS, NA |
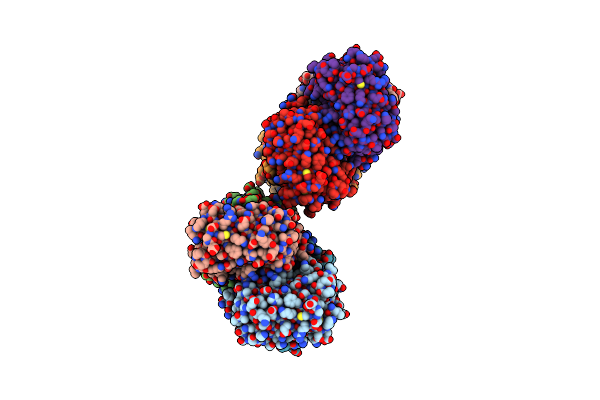 |
Organism: Dendronephthya sp. ssal-2002
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2021-12-08 Classification: FLUORESCENT PROTEIN |
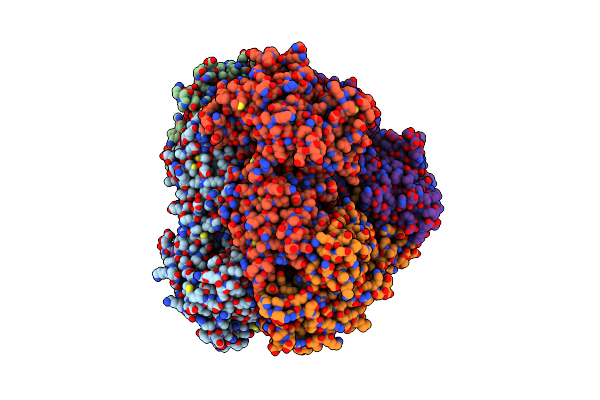 |
Organism: Raoultella planticola
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-05-13 Classification: ISOMERASE Ligands: MN |
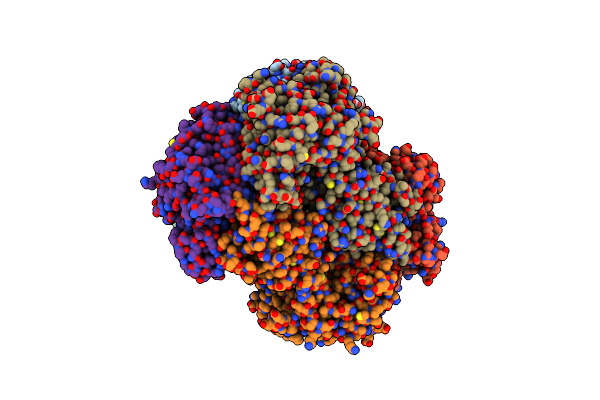 |
Crystal Structure Of The L-Fucose Isomerase Soaked With Mn2+ From Raoultella Sp.
Organism: Raoultella planticola
Method: X-RAY DIFFRACTION Resolution:2.96 Å Release Date: 2020-05-13 Classification: ISOMERASE Ligands: MN |
 |
Organism: Zoanthus sp.
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-01-22 Classification: FLUORESCENT PROTEIN |
 |
Organism: Rhodothermus marinus (strain atcc 43812 / dsm 4252 / r-10)
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2018-05-30 Classification: PROTEIN TRANSPORT |
 |
Crystal Structure Of Glucose Isomerase In Complex With Glycerol In One Metal Binding Mode
Organism: Streptomyces rubiginosus
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2017-09-20 Classification: ISOMERASE Ligands: ACT, MG, GOL |
 |
Crystal Structure Of Glucose Isomerase In Complex With Xylitol Inhibitor In One Metal Binding Mode
Organism: Streptomyces rubiginosus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2017-09-20 Classification: ISOMERASE Ligands: MG, XYL |
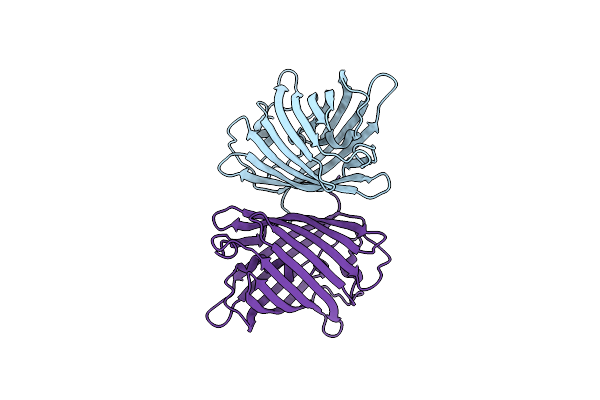 |
Organism: Zoanthus sp.
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2017-09-13 Classification: FLUORESCENT PROTEIN |
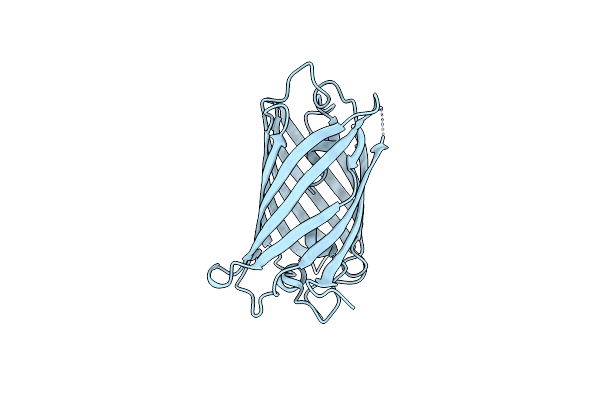 |
Organism: Zoanthus sp.
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-09-13 Classification: FLUORESCENT PROTEIN |
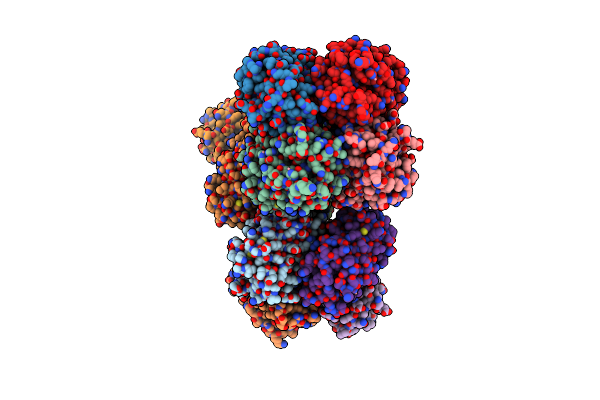 |
Organism: Echinophyllia sp. sc22
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2017-03-15 Classification: LUMINESCENT PROTEIN Ligands: CO |
 |
Organism: Echinophyllia sp. sc22
Method: X-RAY DIFFRACTION Resolution:2.84 Å Release Date: 2017-03-15 Classification: LUMINESCENT PROTEIN Ligands: CU |
 |
Organism: Echinophyllia sp. sc22
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2017-03-15 Classification: LUMINESCENT PROTEIN Ligands: NI |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2012-02-15 Classification: HYDROLASE ACTIVATOR Ligands: SO4 |

