Search Count: 258
 |
Organism: Thermoascus crustaceus
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: HYDROLASE |
 |
Organism: Cellulomonas biazotea
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: HYDROLASE Ligands: GOL |
 |
Crystal Structure Of Beta-Glucosidase Cba3 From Cellulomonas Biazotea In Complex With Glucose
Organism: Cellulomonas biazotea
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: HYDROLASE Ligands: BGC |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Pfr State (I0A).
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL, EDO |
 |
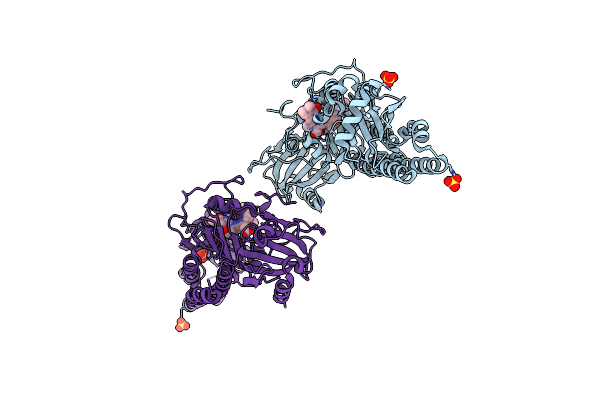
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Pfr State (I0B).
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4 |
 |
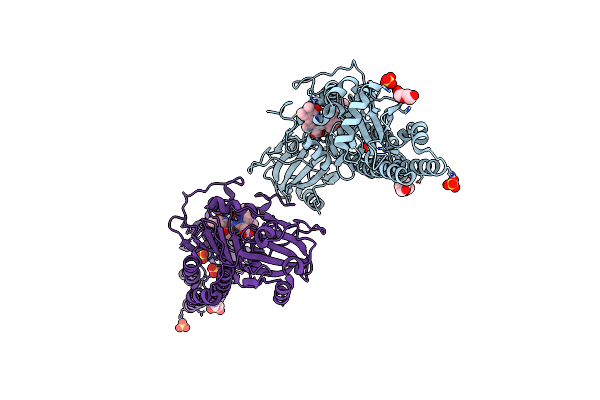
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I1 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4 |
 |
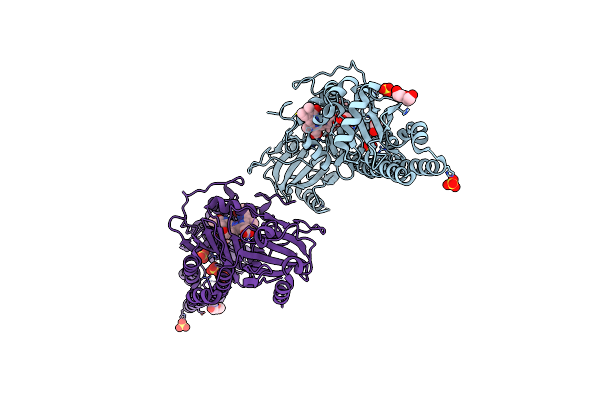
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I2 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, PGE, PEG, CL |
 |
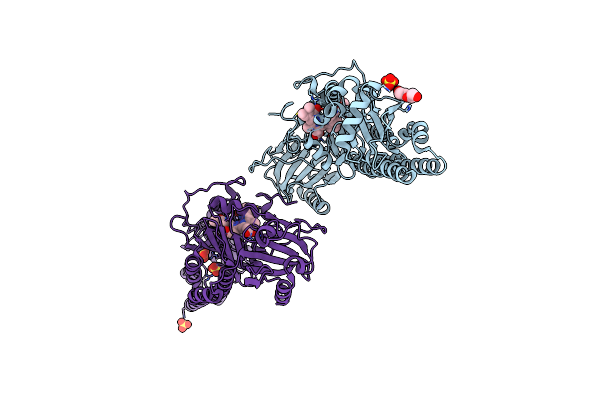
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I3 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, GOL, PEG |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I4 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL, PEG |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I5 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I6 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I7 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, PEG |
 |
Organism: Montipora sp. 20
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-04-23 Classification: FLUORESCENT PROTEIN |
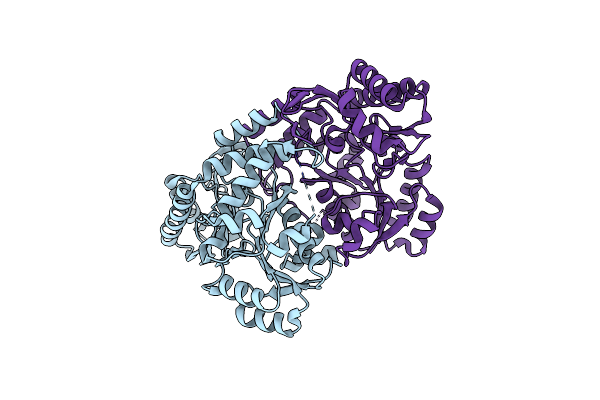 |
Crystal Structure Of Acetylornithine Aminotransferase From Corynebacterium Glutamicum
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-01-17 Classification: TRANSFERASE |
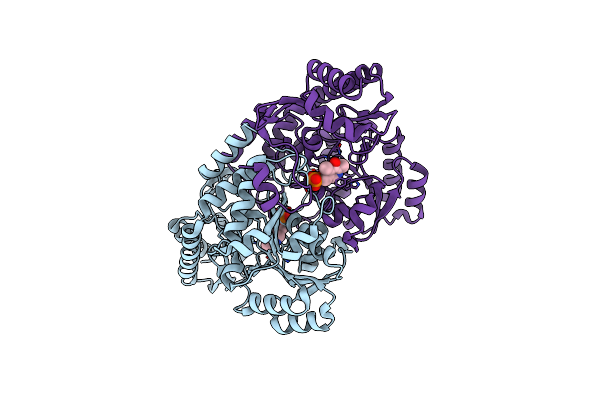 |
Crystal Structure Of Acetylornithine Aminotransferase Complex With Plp From Corynebacterium Glutamicum
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2024-01-17 Classification: TRANSFERASE Ligands: PLP |
 |
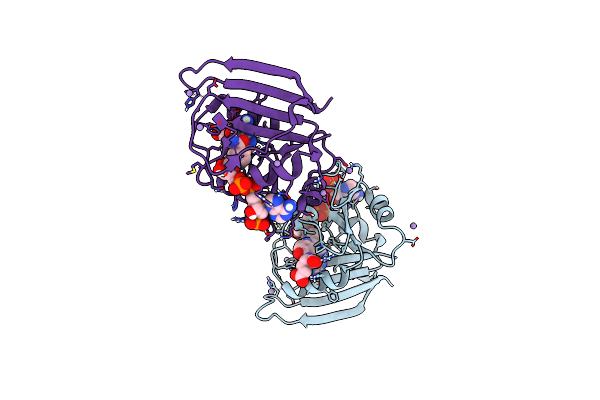
E. Coli Dhfr Complex With Nadp+ And Folate: Ef-X Off Model By Laue Diffraction (No Electric Field)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-01-03 Classification: OXIDOREDUCTASE Ligands: FOL, NAP, MN |
 |
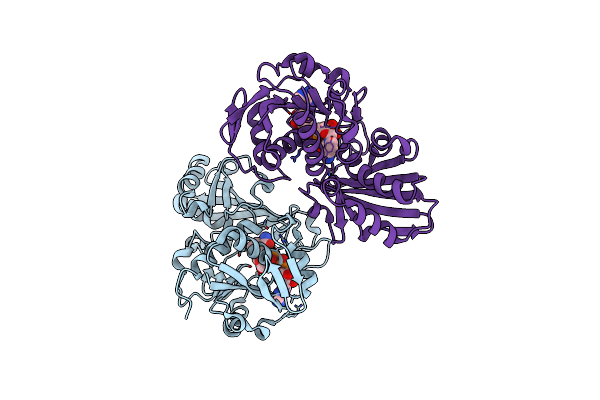
E. Coli Dhfr Complex With Nadp+ And Folate: Ef-X Excited State Model By Laue Diffraction (Electric Field Along B Axis; 8-Fold Extrapolation Of Structure Factor Differences)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-01-03 Classification: OXIDOREDUCTASE Ligands: FOL, NAP, MN |
 |
Crystal Structure Of Glyceraldehyde-3-Phosphate Dehydrogenase From Corynebacterium Glutamicum Atcc13032 In Complex With Nad
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2023-12-06 Classification: STRUCTURAL PROTEIN Ligands: NAD |
 |
Crystal Structure Of Glyceraldehyde-3-Phosphate Dehydrogenase From Corynebacterium Glutamicum Atcc13032 In Complex With Nad And G3P
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2023-12-06 Classification: STRUCTURAL PROTEIN Ligands: NAD, G3H, GOL, EDO, PG4 |
 |
Crystal Structure Of Glyceraldehyde-3-Phosphate Dehydrogenase From Corynebacterium Glutamicum Atcc13032 (L36S/T37K) In Complex With Nad
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2023-12-06 Classification: STRUCTURAL PROTEIN Ligands: CS, NAD, EDO, GOL, PO4 |

