Search Count: 51
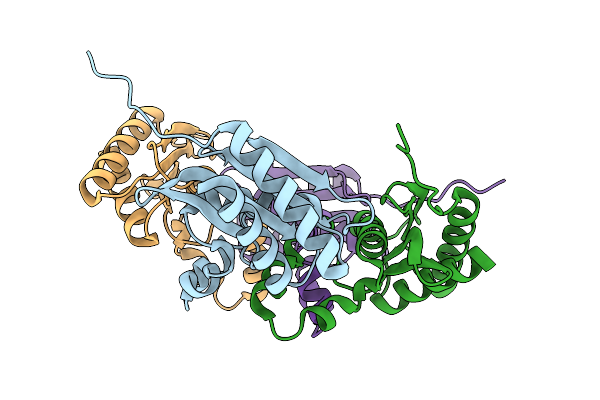 |
Organism: Pseudomonas sp. mc1
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: UNKNOWN FUNCTION |
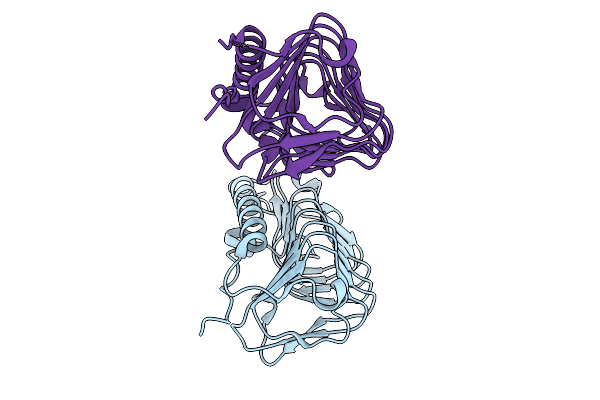 |
Crystal Structure Of Ice Binding Protein From Candidatus Cryosericum Odellii Smc5
Organism: Candidatus cryosericum odellii
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: RECOMBINATION |
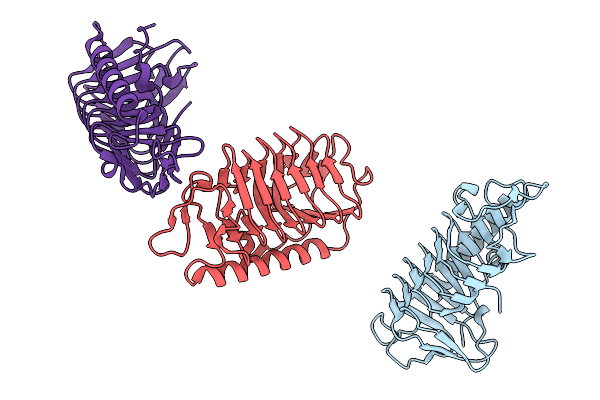 |
Crystal Structure Of Ice Binding Protein From Candidatus Cryosericum Odellii Smc5
Organism: Candidatus cryosericum odellii
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: RECOMBINATION |
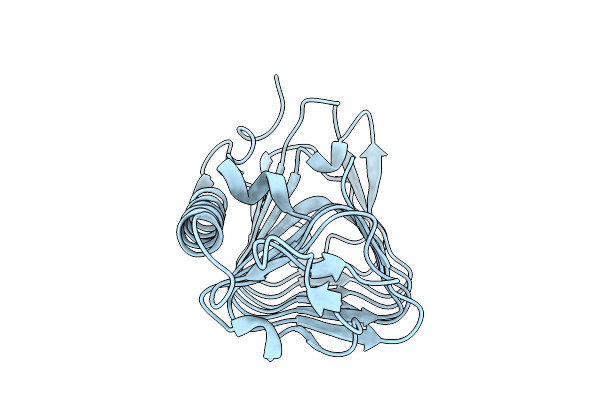 |
Crystal Structure Of An Ice-Binding Protein From Candidatus Cryosericum Odellii
Organism: Candidatus cryosericum odellii
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: PROTEIN BINDING |
 |
Crystal Structure Of Engineered Ice-Binding Protein (M74I And A97V) From Candidatus Cryosericum Odellii Strain Smc5_169
Organism: Candidatus cryosericum odellii
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: PROTEIN BINDING |
 |
Organism: Exiguobacterium antarcticum b7
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2023-09-20 Classification: HYDROLASE |
 |
Organism: Lactococcus garvieae subsp. garvieae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-06-07 Classification: HYDROLASE Ligands: CL, ACT |
 |
Organism: Lactococcus garvieae subsp. garvieae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-06-07 Classification: HYDROLASE |
 |
Organism: Lactobacillus acidophilus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-05-17 Classification: HYDROLASE |
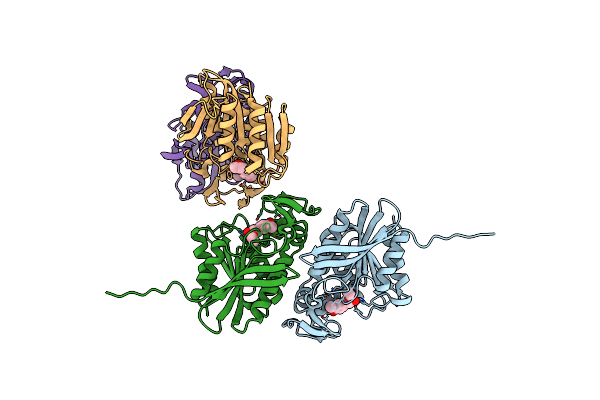 |
Organism: Lactobacillus acidophilus
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2023-05-17 Classification: HYDROLASE Ligands: ZYC |
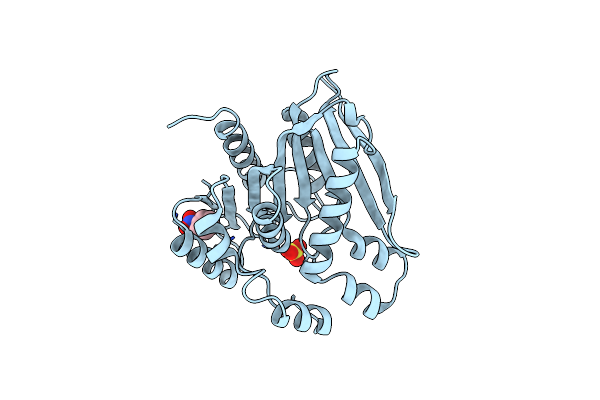 |
Organism: Paenibacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-08-31 Classification: HYDROLASE Ligands: SO4, NBZ |
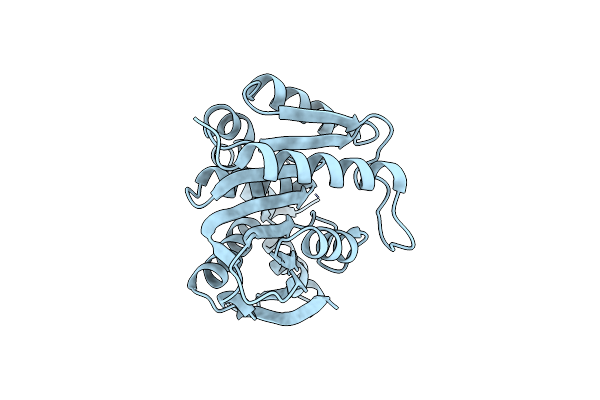 |
Organism: Paenibacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-08-31 Classification: HYDROLASE |
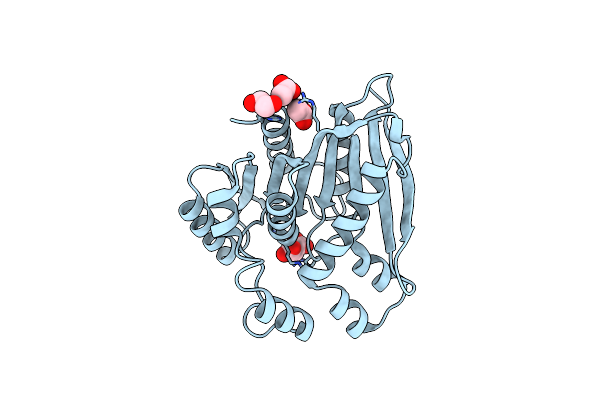 |
Organism: Paenibacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-08-31 Classification: HYDROLASE Ligands: MLA, EDO, ACT |
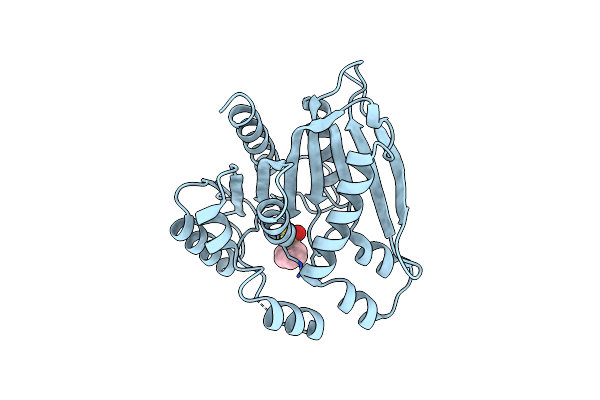 |
Crystal Structure Of Psest3 Complexed With Phenylmethylsulfonyl Fluoride (Pmsf)
Organism: Paenibacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2022-08-31 Classification: HYDROLASE Ligands: PMS |
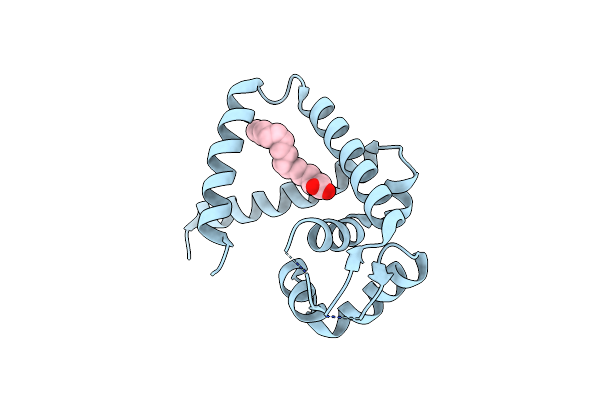 |
Crystal Structure Of A Marr Family Protein In Complex With A Lipid-Like Effector Molecule From The Psychrophilic Bacterium Paenisporosarcina Sp. Tg-14
Organism: Paenisporosarcina sp. tg-14
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-11-24 Classification: DNA BINDING PROTEIN Ligands: PLM |
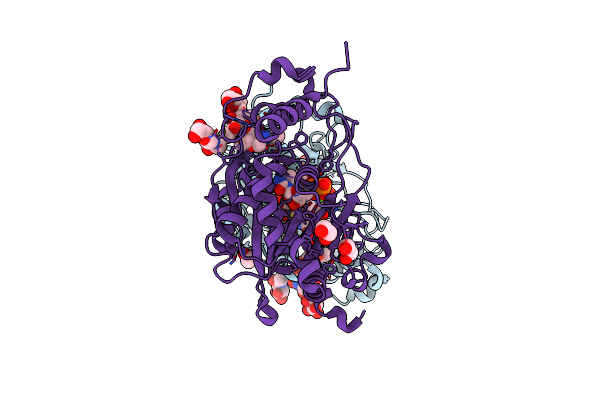 |
Human Poly-N-Acetyl-Lactosamine Synthase Structure Demonstrates A Modular Assembly Of Catalytic Subsites For Gt-A Glycosyltransferases
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2020-12-02 Classification: TRANSFERASE Ligands: MG, UDP, NAG, EDO, EPE |
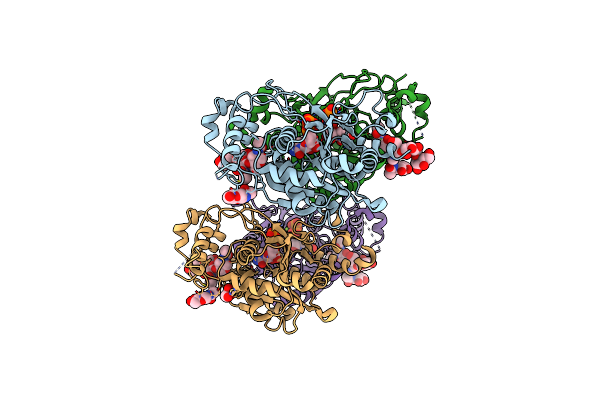 |
Human Poly-N-Acetyl-Lactosamine Synthase Structure Demonstrates A Modular Assembly Of Catalytic Subsites For Gt-A Glycosyltransferases
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2020-12-02 Classification: TRANSFERASE Ligands: MG, UDP, NAG, EDO, CL |
 |
Human Poly-N-Acetyl-Lactosamine Synthase Structure Demonstrates A Modular Assembly Of Catalytic Subsites For Gt-A Glycosyltransferases
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-12-02 Classification: TRANSFERASE Ligands: MG, UDP, NAG, EDO, GOL |
 |
Monodehydroascorbate Reductase, Mdhar, From Antarctic Hairgrass Deschampsia Antarctica
Organism: Deschampsia antarctica
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-08-26 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2020-05-20 Classification: TRANSFERASE Ligands: ADP, SO4, CL, EDO, DMS |

