Search Count: 314
 |
Solution Structure Of Holo Acyl Carrier Protein 2 (Apef) Of Aryl Polyene Biosynthesis From Acinetobacter Baumannii
Organism: Acinetobacter baumannii
Method: SOLUTION NMR Release Date: 2025-11-26 Classification: BIOSYNTHETIC PROTEIN |
 |
Solution Structure Of Holo Acyl Carrier Protein 1 (Apee) Of Aryl Polyene Biosynthesis From Acinetobacter Baumannii
Organism: Acinetobacter baumannii
Method: SOLUTION NMR Release Date: 2025-11-26 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Aequorea
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: FLUORESCENT PROTEIN Ligands: CIT, 1PE, SO4 |
 |
Organism: Mus musculus, Synthetic constrcut
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TRANSFERASE |
 |
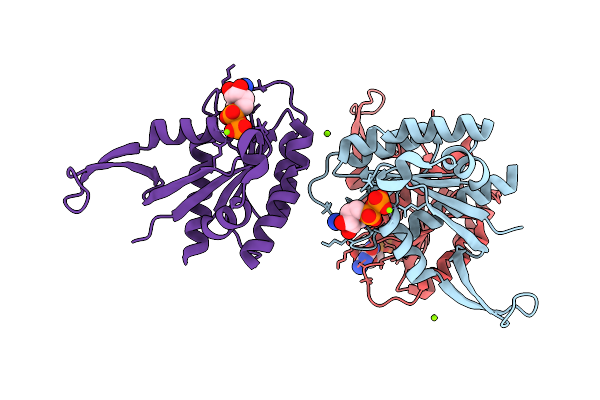
Crystal Structure Of Gdp-Bound Kras G12D/M67R: Suppressing G12D Oncogenicity Via Second-Site M67R Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: ONCOPROTEIN Ligands: GDP, MG |
 |
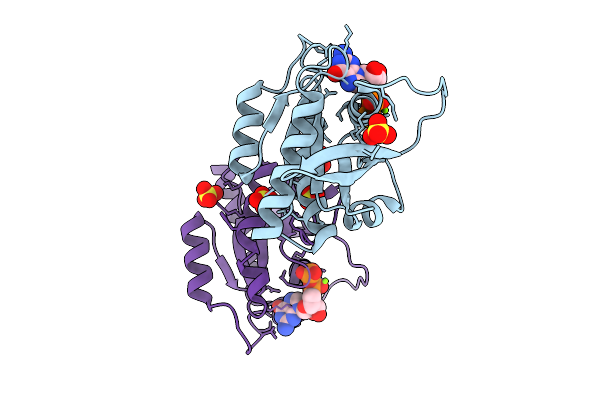
Crystal Structure Of Gdp-Bound Kras G12D/I55E: Suppressing G12D Oncogenicity Via Second-Site I55E Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: ONCOPROTEIN Ligands: GDP, MG |
 |
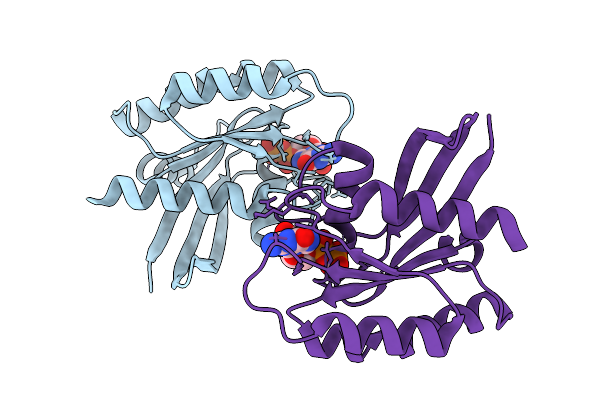
Crystal Structure Of Gdp-Bound Kras G12D/F28K: Suppressing G12D Oncogenicity Via Second-Site F28K Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: ONCOPROTEIN Ligands: GDP, MG, SO4 |
 |
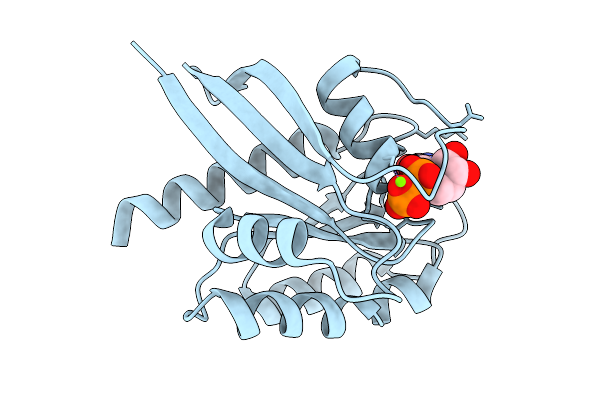
Crystal Structure Of Gdp-Bound Kras G12D/P34R: Suppressing G12D Oncogenicity Via Second-Site P34R Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: ONCOPROTEIN Ligands: MG, GDP |
 |
Crystal Structure Of Gdp-Bound Kras G12D/R41Q: Suppressing G12D Oncogenicity Via Second-Site R41Q Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: ONCOPROTEIN Ligands: GDP, MG |
 |
Crystal Structure Of Gdp-Bound Kras G12D/V45E: Suppressing G12D Oncogenicity Via Second-Site V45E Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: ONCOPROTEIN Ligands: GDP, MG |
 |
Crystal Structure Of Gdp-Bound Kras G12D/D54R: Suppressing G12D Oncogenicity Via Second-Site D54R Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: ONCOPROTEIN Ligands: GDP, MG, SO4 |
 |
Crystal Structure Of Gdp-Bound Kras G12D/G60R: Suppressing G12D Oncogenicity Via Second-Site G60R Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: ONCOPROTEIN Ligands: MG, GDP |
 |
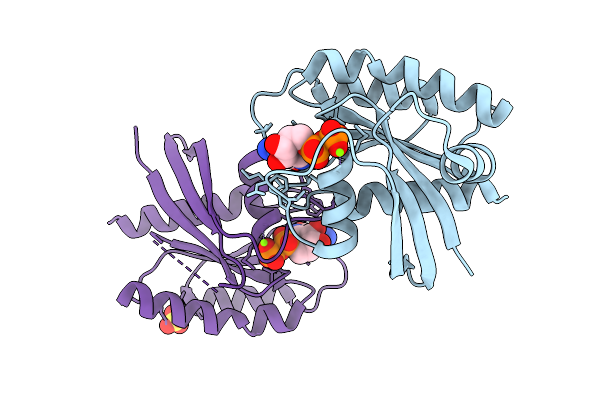
Crystal Structure Of Gdp-Bound Kras G12D/V103Y: Suppressing G12D Oncogenicity Via Second-Site V103Y Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: ONCOPROTEIN Ligands: MG, GDP |
 |
Crystal Structure Of Gdp-Bound Kras G12D/E62Q: Suppressing G12D Oncogenicity Via Second-Site E62Q Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: ONCOPROTEIN Ligands: MG, GDP, SO4 |
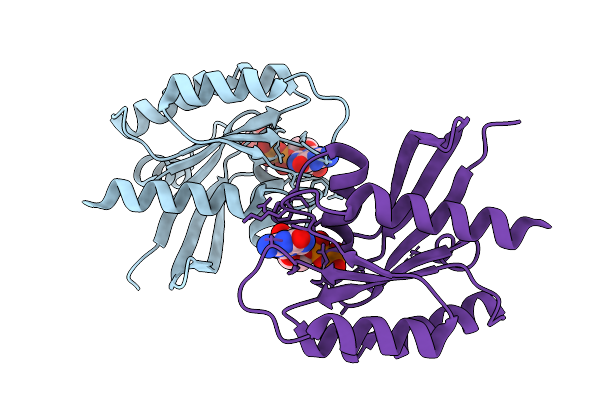 |
Crystal Structure Of Gdp-Bound Kras E3K/G12D: Suppressing G12D Oncogenicity Via Second-Site E3K Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: ONCOPROTEIN Ligands: MG, GDP, GOL |
 |
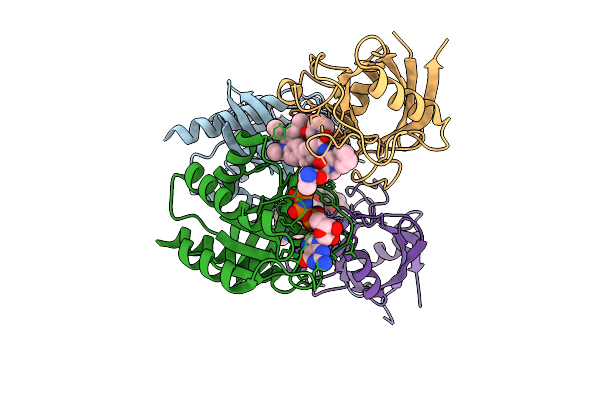
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: HYDROLASE Ligands: GNP, MG, CL, A1AZV |
 |
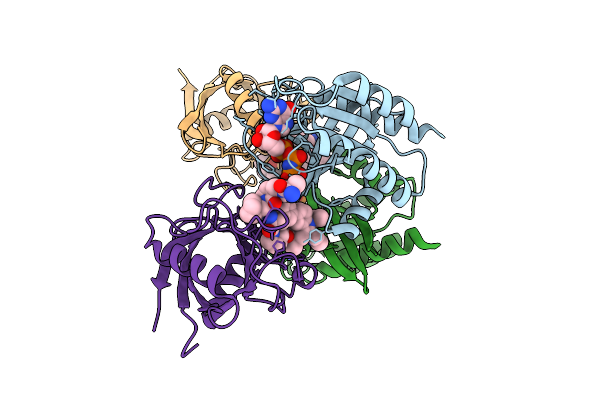
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: GNP, MG, A1AZX, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: GNP, MG, CL, A1AZY |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: GNP, MG, A1AZW |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: GNP, MG, CL, A1AZZ |

