Search Count: 59
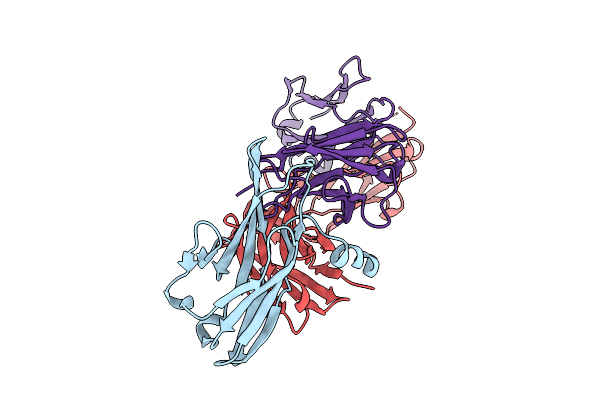 |
Crystal Structure Of Pilra In Complex With Fab Portion Of Antagonist Antibody
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: SIGNALING PROTEIN |
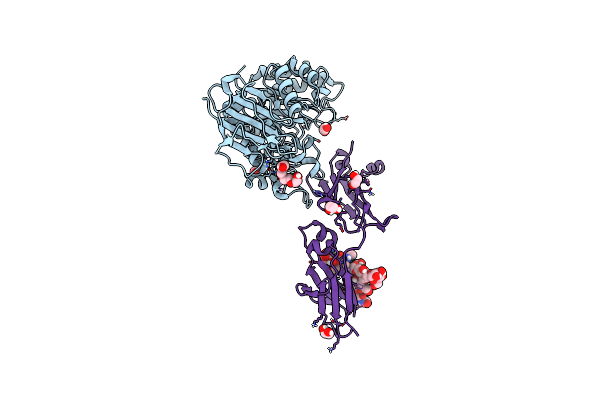 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2023-10-18 Classification: TRANSPORT PROTEIN Ligands: EDO, PG4 |
 |
Crystal Structure Of An Alpha/Beta-Hydrolase Enzyme From Chloroflexus Sp. Ms-G (202)
Organism: Chloroflexus sp. ms-g
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2022-12-28 Classification: HYDROLASE |
 |
Crystal Structure Of An Alpha/Beta-Hydrolase Enzyme From Candidatus Kryptobacter Tengchongensis (306)
Organism: Candidatus kryptobacter tengchongensis
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: PO4 |
 |
Crystal Structure Of A Cutinase Enzyme From Marinactinospora Thermotolerans Dsm45154 (606)
Organism: Marinactinospora thermotolerans
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2022-12-28 Classification: HYDROLASE |
 |
Organism: Saccharopolyspora flava
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: PG4 |
 |
Organism: Thermobifida fusca
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: PEG |
 |
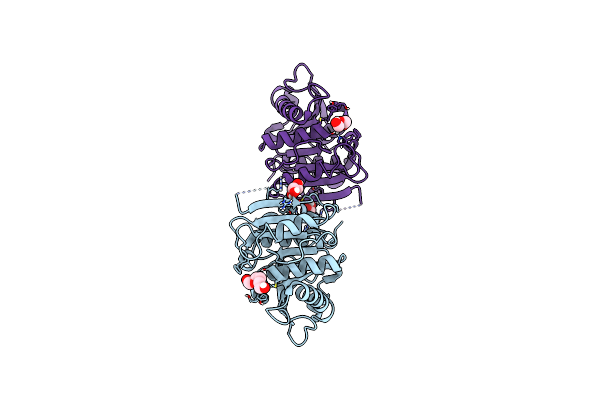
Organism: Thermobifida fusca
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: PG4 |
 |
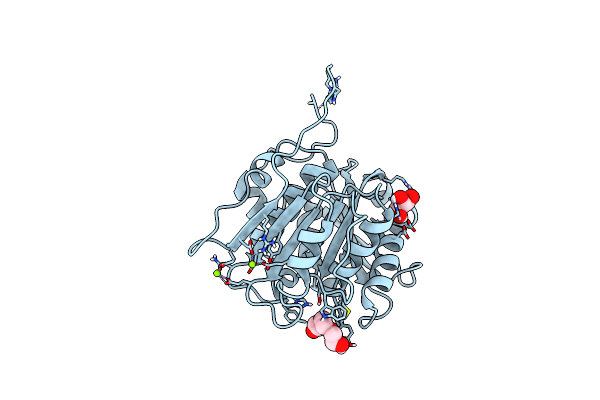
Organism: Thermobifida fusca yx
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: GOL, PG4, PEG |
 |
Crystal Structure Of A Cutinase Enzyme From Thermobifida Cellulosilytica Tb100 (711)
Organism: Thermobifida cellulosilytica tb100
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: GOL, PG4, MG |
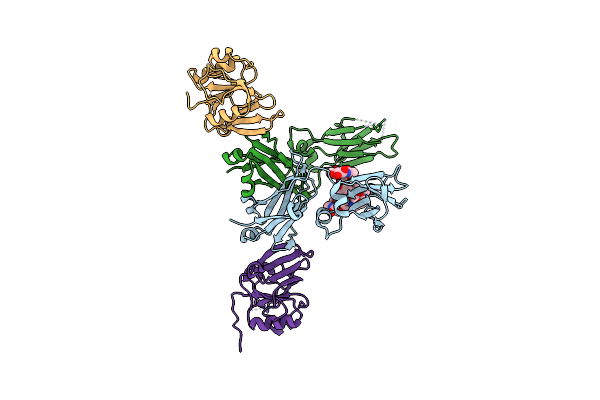 |
Brain Delivery Of Therapeutic Proteins Using An Fc Fragment Blood-Brain Barrier Transport Vehicle In Mice And Monkeys
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.38 Å Release Date: 2020-06-10 Classification: PROTEIN TRANSPORT |
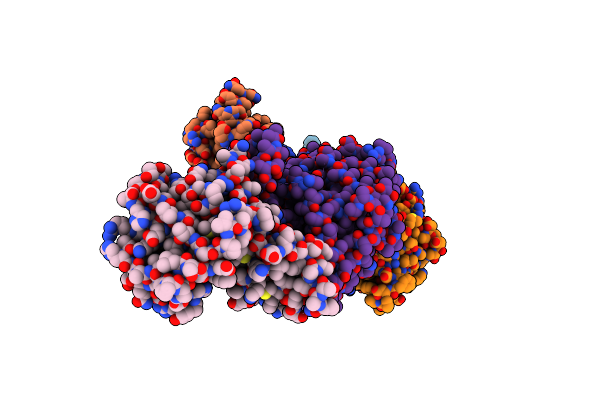 |
Cryo-Em Structure Of The Lysosomal Folliculin Complex (Flcn-Fnip2-Raga-Ragc-Ragulator)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.60 Å Release Date: 2019-11-06 Classification: signaling protein/inhibitor Ligands: GDP, L8S |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2017-11-08 Classification: SIGNALING PROTEIN |
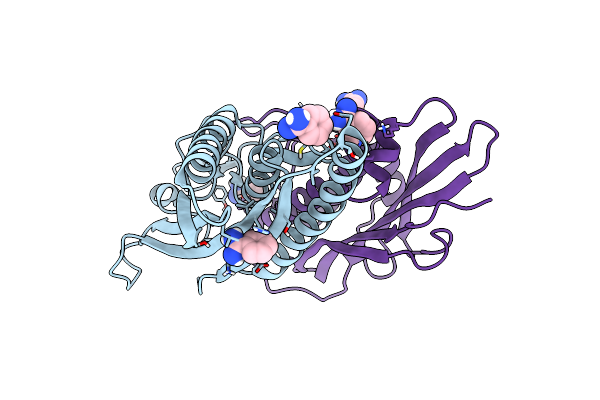 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2015-10-14 Classification: PROTEIN BINDING Ligands: BEN |
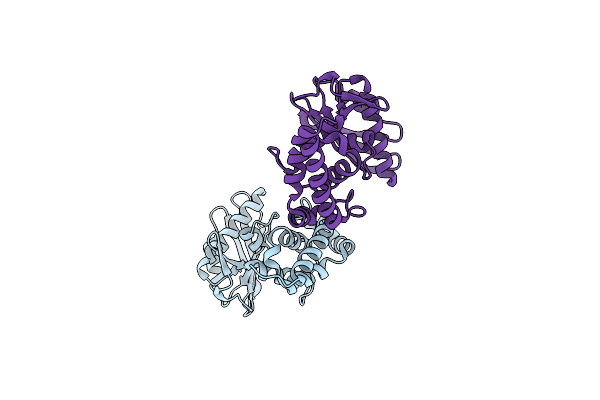 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2014-04-09 Classification: TRANSFERASE |
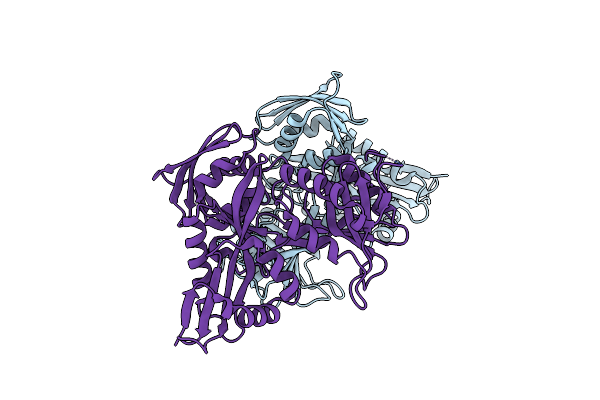 |
Mycobacterium Tuberculosis Eis Protein Initiates Modulation Of Host Immune Responses By Acetylation Of Dusp16/Mkp-7
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-07-25 Classification: TRANSFERASE |
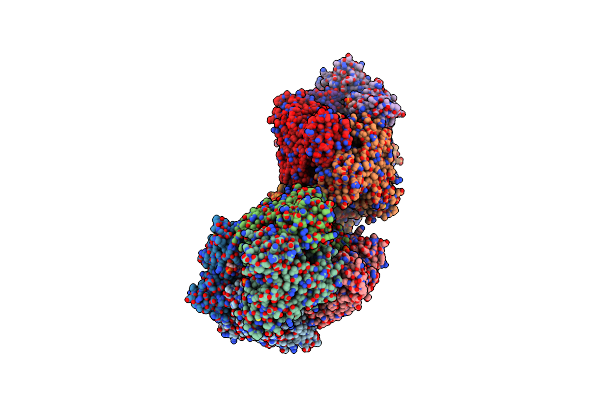 |
Crystal Structure Of Enhanced Intracellular Survival (Eis) Protein From Mycobacterium Tuberculosis With Acetyl Coa
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2012-05-23 Classification: TRANSFERASE Ligands: ACO |
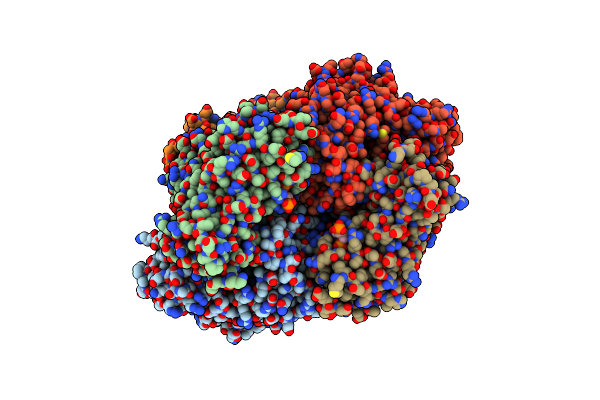 |
Mycobacterium Tuberculosis Eis Protein Initiates Modulation Of Host Immune Responses By Acetylation Of Dusp16/Mkp-7
Organism: Mycobacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2012-05-23 Classification: TRANSFERASE Ligands: COA |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-05-23 Classification: TRANSFERASE |
 |
Structural And Functional Insights Of Directly Targeting Pin1 By Epigallocatechin-3-Gallate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2011-08-17 Classification: ISOMERASE Ligands: SO4, PE4, KDH |

