Search Count: 59
 |
Organism: Streptomyces lasalocidi
Method: X-RAY DIFFRACTION Release Date: 2025-01-15 Classification: FLAVOPROTEIN Ligands: DMS, A1LWM, FAD, CL |
 |
Organism: Streptomyces lasalocidi
Method: X-RAY DIFFRACTION Release Date: 2025-01-15 Classification: FLAVOPROTEIN Ligands: FAD, LSB, CL |
 |
Organism: Streptomyces lasalocidi
Method: X-RAY DIFFRACTION Release Date: 2024-12-18 Classification: FLAVOPROTEIN Ligands: FAD, CL |
 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-11-08 Classification: OXIDOREDUCTASE Ligands: FAD, CL |
 |
Crystal Structure Of The Multidrug Efflux Transporter Bpeb From Burkholderia Pseudomallei
Organism: Burkholderia pseudomallei k96243
Method: X-RAY DIFFRACTION Resolution:2.94 Å Release Date: 2023-07-19 Classification: MEMBRANE PROTEIN Ligands: UMQ, PG4 |
 |
Crystal Structure Of The Multidrug Effulx Transporter Bpef From Burkholderia Pseudomallei.
Organism: Burkholderia pseudomallei k96243
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2023-07-19 Classification: MEMBRANE PROTEIN Ligands: LMT |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-01-11 Classification: PLANT PROTEIN Ligands: CA, EDO |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2023-01-11 Classification: PLANT PROTEIN Ligands: T83, CA, EDO, PG4, ACT |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2023-01-11 Classification: PLANT PROTEIN Ligands: 07L, CA, ACT |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2023-01-11 Classification: PLANT PROTEIN Ligands: COA, GOL, CA |
 |
Organism: Streptomyces lasalocidi
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2022-10-26 Classification: ANTIBIOTIC Ligands: ADP, ZN, CL, MG |
 |
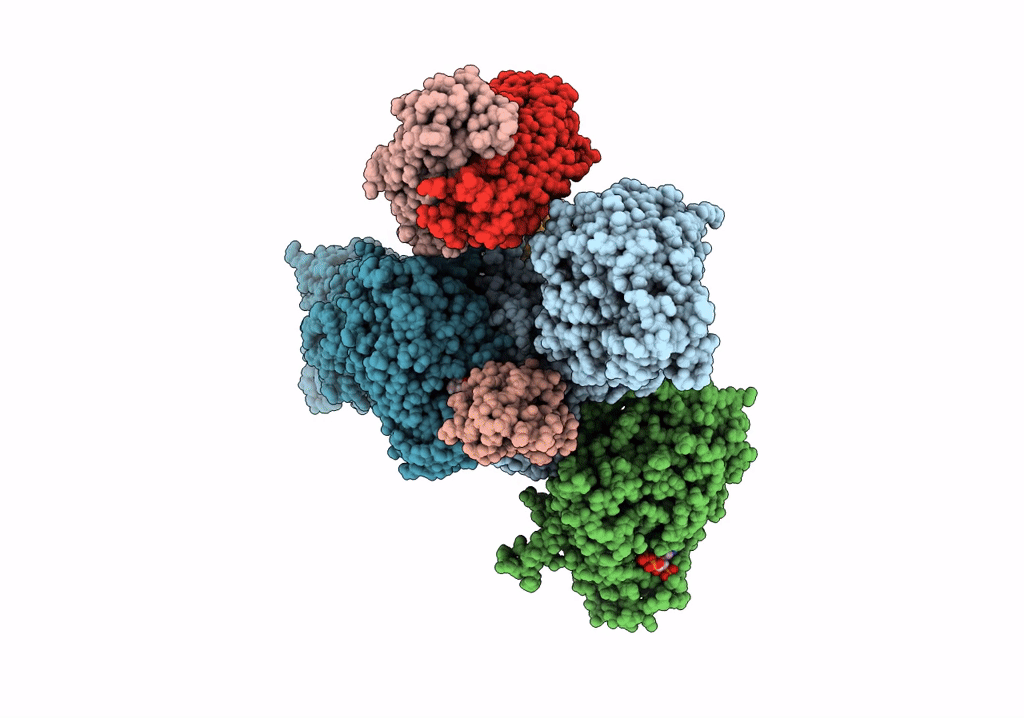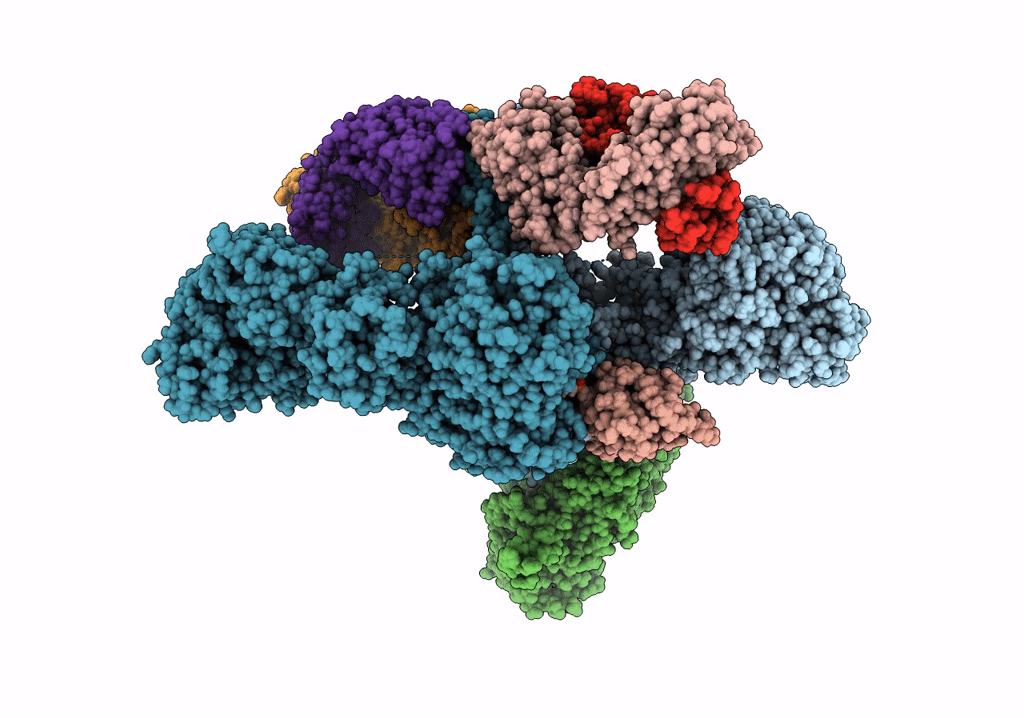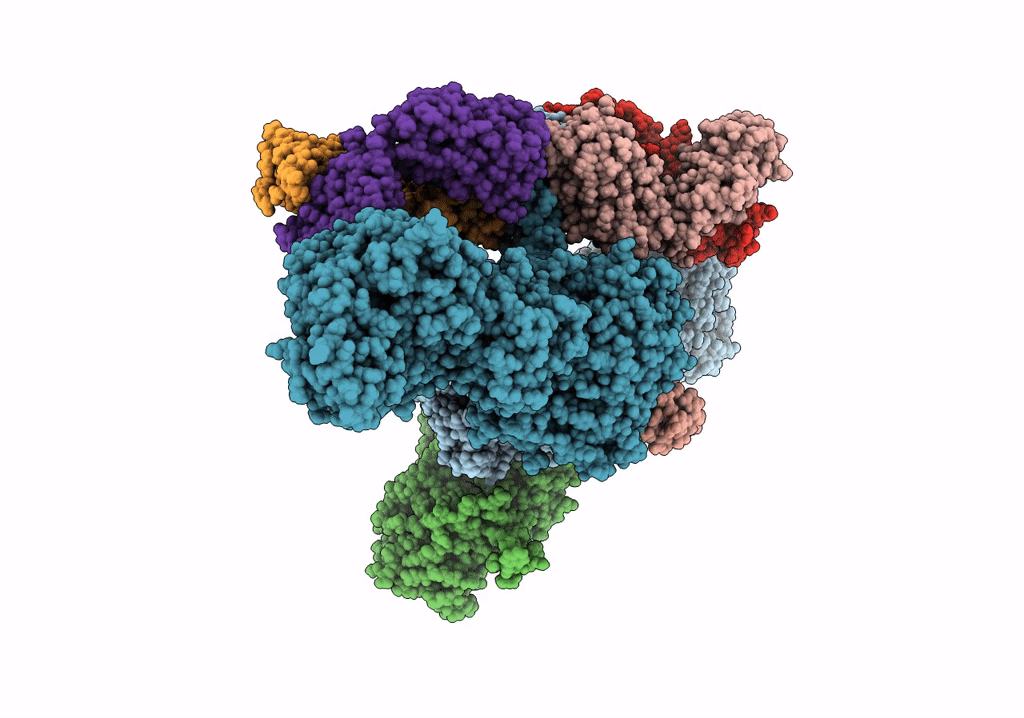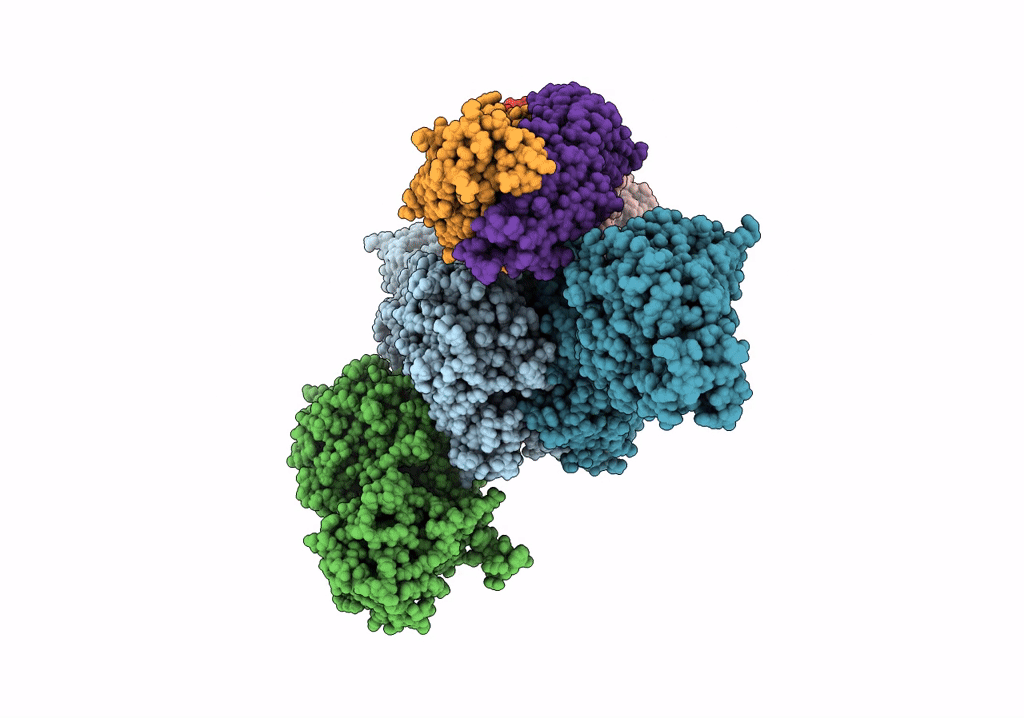
Crystal Structure Of Modular Polyketide Synthase Apo-Lsd14 From The Lasalocid Biosynthesis Pathway, Trapped In The Transacylation Step
Organism: Streptomyces lasalocidi
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2021-11-03 Classification: BIOSYNTHETIC PROTEIN |
 |
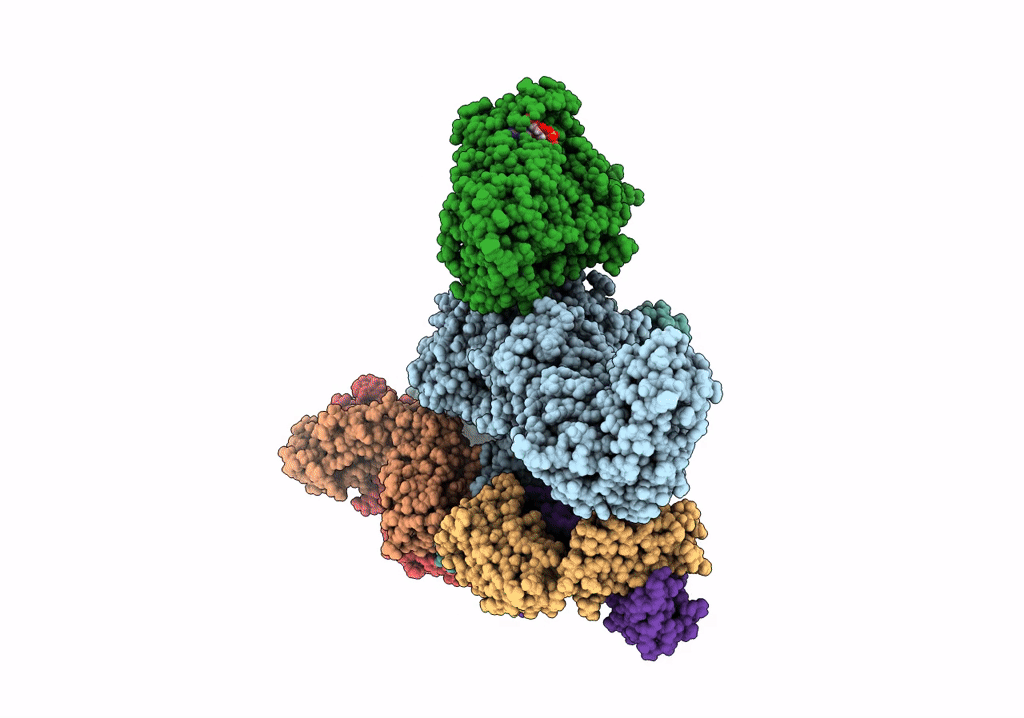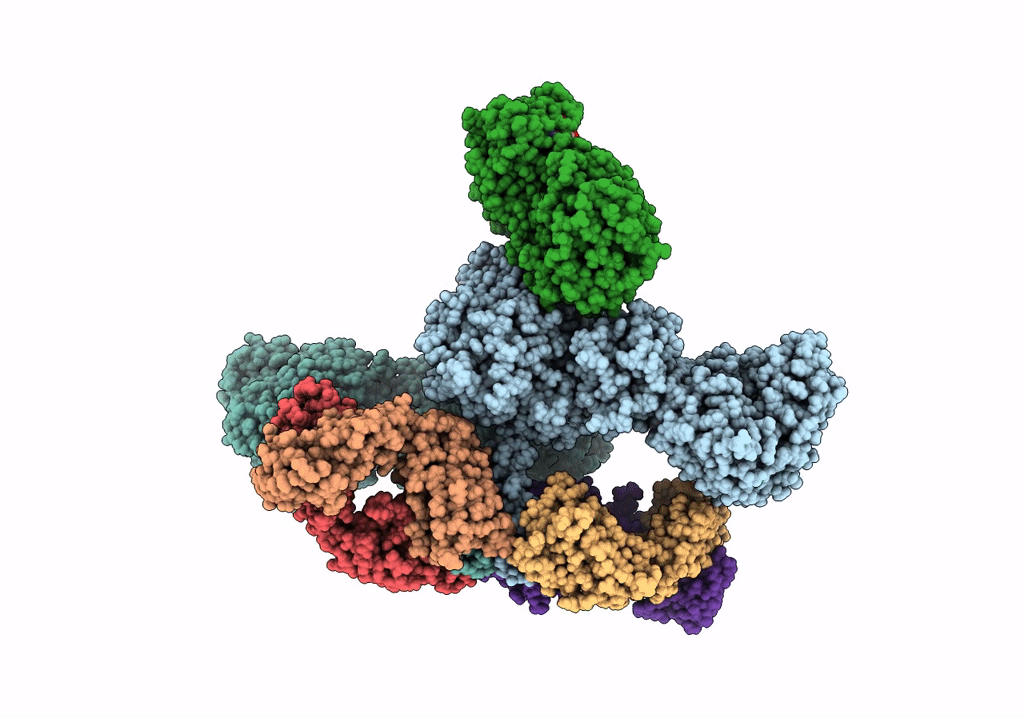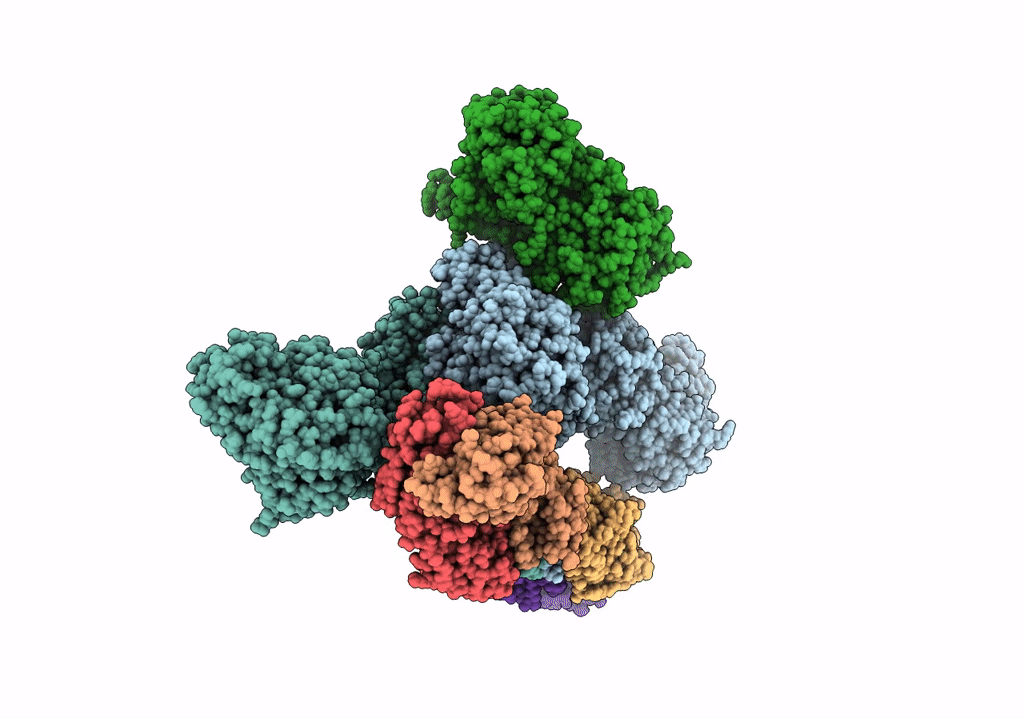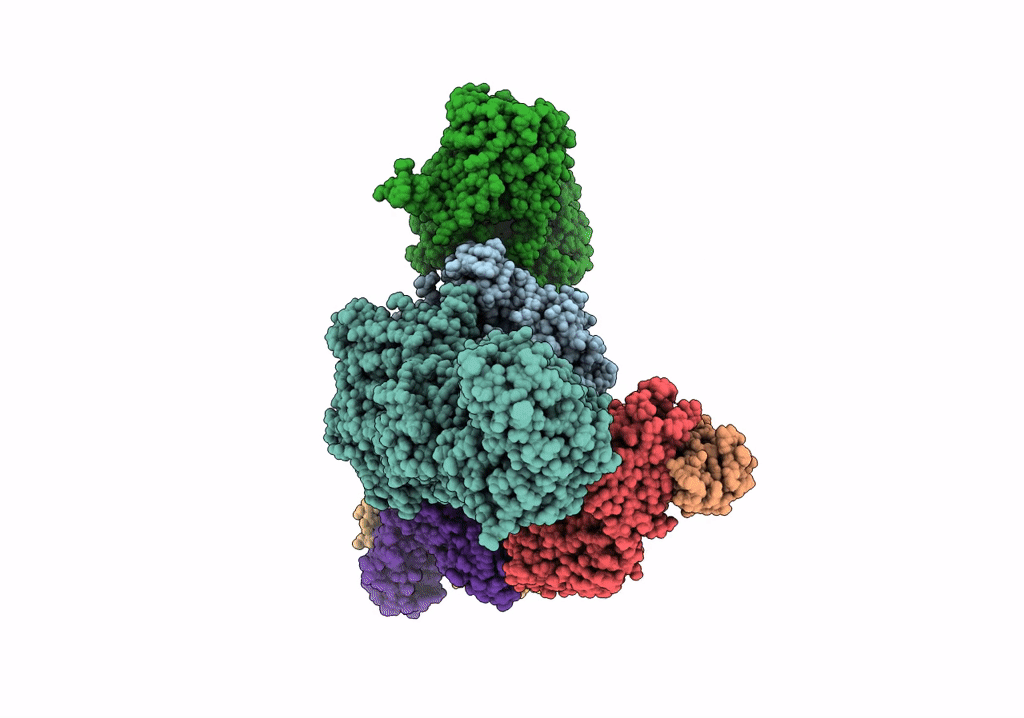
Cryoem Structure Of Modular Pks Holo-Lsd14 Stalled At The Condensation Step And Bound To Antibody Fragment 1B2, Composite Structure
Organism: Saccharopolyspora erythraea, Streptomyces lasalocidi, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-11-03 Classification: BIOSYNTHETIC PROTEIN Ligands: ATR, PNS |
 |
Cryoem Structure Of Modular Pks Holo-Lsd14 Bound To Antibody Fragment 1B2, Composite Structure
Organism: Saccharopolyspora erythraea, Streptomyces lasalocidi, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-11-03 Classification: BIOSYNTHETIC PROTEIN Ligands: ATR |
 |
Organism: Deinococcus metallilatus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-08-25 Classification: LIGASE |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2021-03-03 Classification: HYDROLASE Ligands: F4X, MG, NA, DMS, GOL |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2021-03-03 Classification: HYDROLASE Ligands: F6L, MG, NA, DMS, GOL |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2020-07-08 Classification: HYDROLASE Ligands: DVL, NA, DMS, MG, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.07 Å Release Date: 2017-11-22 Classification: HYDROLASE/PROTEIN BINDING Ligands: NAG, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2017-04-05 Classification: IMMUNE SYSTEM |

