Search Count: 515
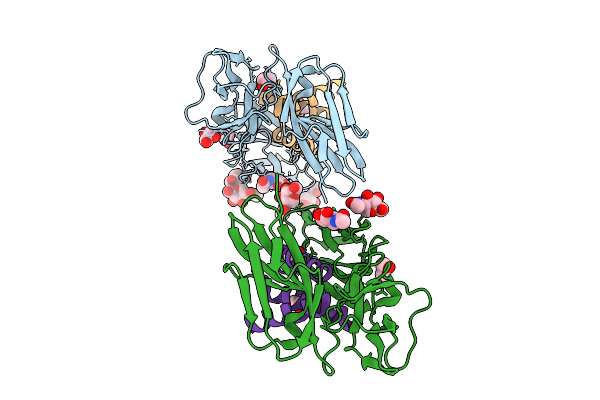 |
Kit123-Kitbp Complex (Domains D1-3 Of The Human Receptor Tyrosine Kinase Kit Complexed With The De Novo Designed Minibinder Kitbp)
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2026-01-21 Classification: PROTEIN BINDING Ligands: NAG, GOL, PEG, EDO |
 |
Organism: Synthetic construct, Homo sapiens, Human t-cell leukemia virus type i
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2026-01-14 Classification: IMMUNE SYSTEM Ligands: NAG, SO4 |
 |
Organism: Mus musculus, Mammarenavirus choriomeningitidis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-12-31 Classification: IMMUNE SYSTEM |
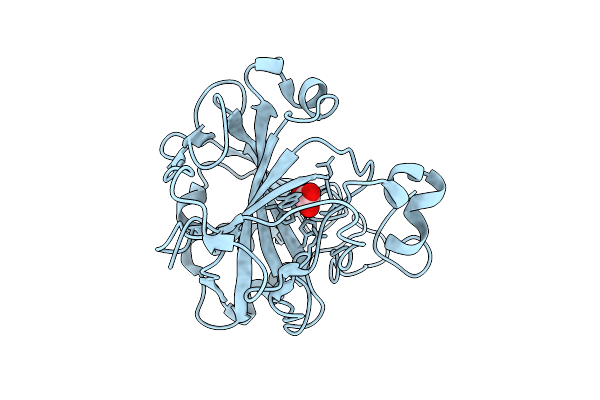 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2025-10-22 Classification: METAL BINDING PROTEIN Ligands: CO, BCT |
 |
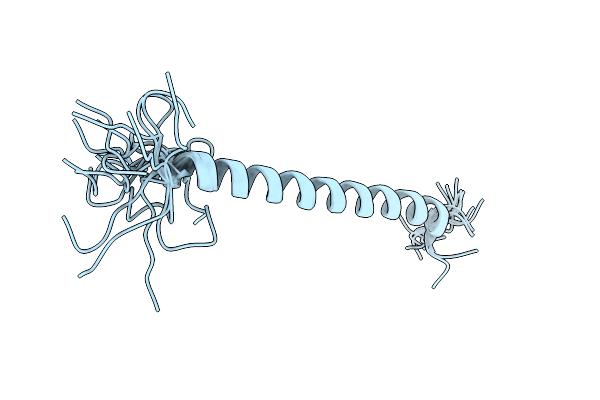
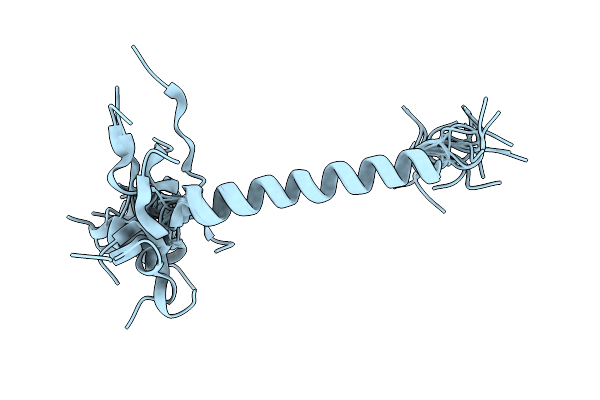
Structure Of A De Novo Designed Interleukin-21 Mimetic Complex With Il-21R And Il-2Rg
Organism: Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2025-09-24 Classification: DE NOVO PROTEIN/IMMUNE SYSTEM Ligands: NAG, EDO |
 |
 |
 |
Room Temperature Structure Of Kr2 Rhodopsin In Pentameric Form At 95% Relative Humidity
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2025-07-16 Classification: MEMBRANE PROTEIN Ligands: RET, NA, OLC, LFA |
 |
Room-Temperature Structure Of Kr2 Rhodopsin In Pentameric Form At 85% Relative Humidity
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2025-07-16 Classification: MEMBRANE PROTEIN Ligands: RET, NA, OLC, LFA |
 |
Hen Egg-White Lysozyme Structure Embedded In Lcp Medium At 95% Relative Humidity
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-07-16 Classification: HYDROLASE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-07-16 Classification: HYDROLASE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-07-16 Classification: HYDROLASE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-07-16 Classification: HYDROLASE |
 |
Hen Egg-White Lysozyme Structure Collected At Euxfel Spb/Sfx With Hve Injection Method
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-07-16 Classification: HYDROLASE |
 |
Hen Egg-White Lysozyme Structure Embedded In Lcp Medium At 95% Relative Humidity
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-07-16 Classification: HYDROLASE |
 |
Hen Egg-White Lysozyme Structure Embedded In Lcp Medium At 85% Relative Humidity
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-07-16 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2025-07-16 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.23 Å Release Date: 2025-07-16 Classification: METAL BINDING PROTEIN, LYASE Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2025-07-16 Classification: METAL BINDING PROTEIN, LYASE Ligands: ZN, CO2 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2025-07-16 Classification: LYASE Ligands: ZN, BCT |

