Search Count: 33
 |
Organism: Homo sapiens, Bos taurus, Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2022-02-09 Classification: MEMBRANE PROTEIN Ligands: NKP |
 |
Organism: Homo sapiens, Bos taurus, Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2022-02-09 Classification: MEMBRANE PROTEIN Ligands: NKP |
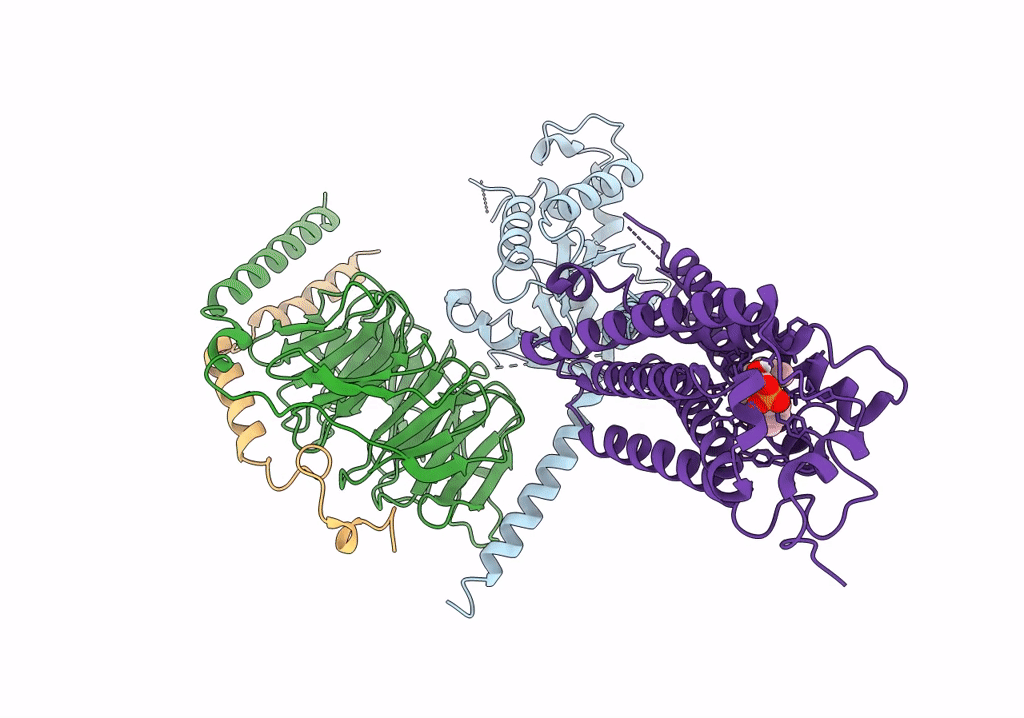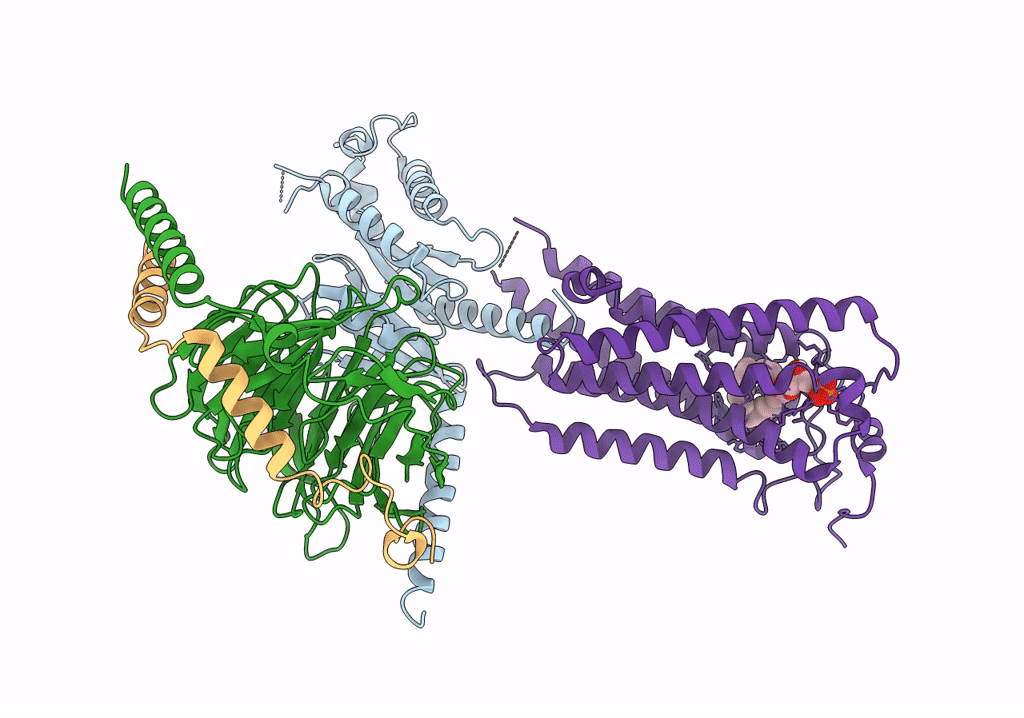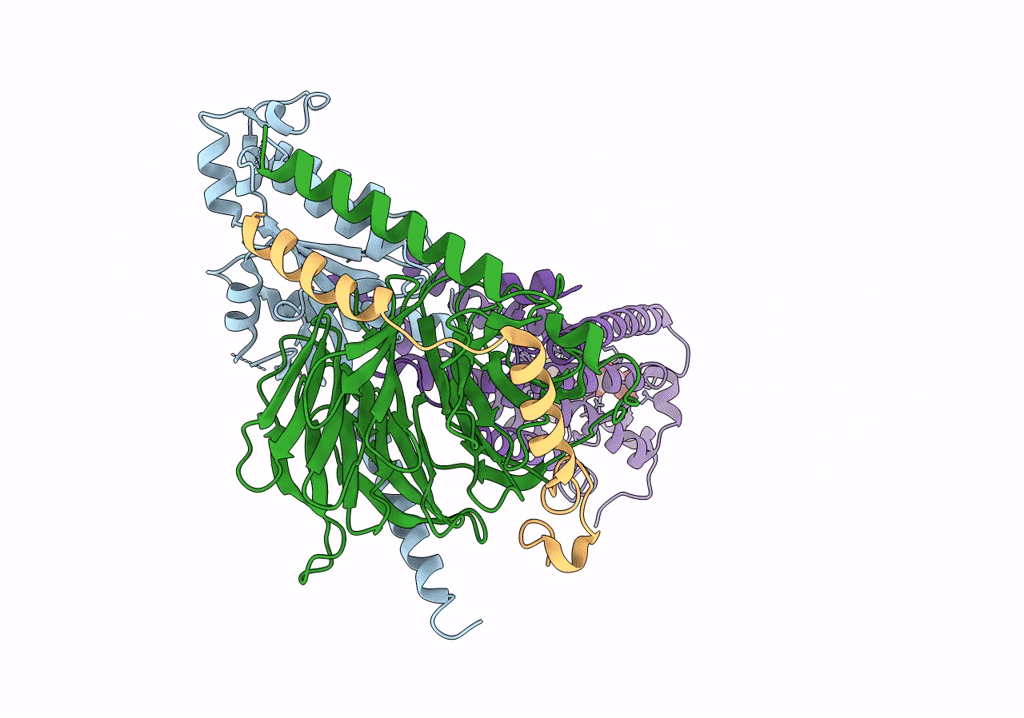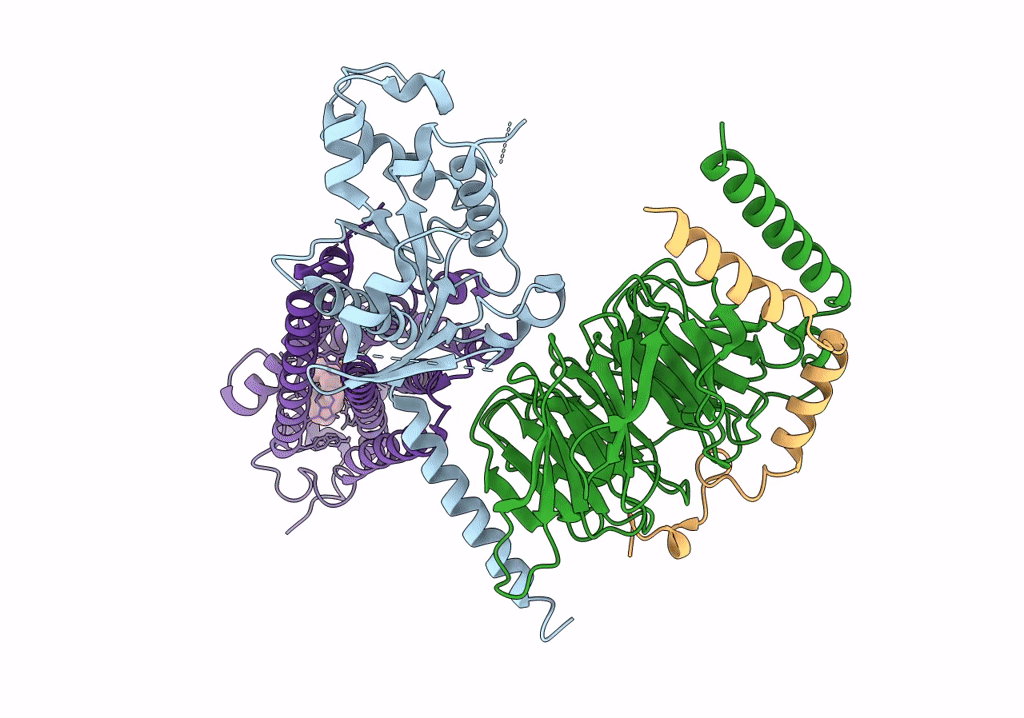 |
Organism: Homo sapiens, Bos taurus, Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2022-02-09 Classification: MEMBRANE PROTEIN Ligands: NKP |
 |
Organism: Bos taurus, Rattus norvegicus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-02-09 Classification: MEMBRANE PROTEIN Ligands: S1P, NAG |
 |
Organism: Bos taurus, Rattus norvegicus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-02-09 Classification: MEMBRANE PROTEIN Ligands: NAG, J8C |
 |
Crystal Structure Of The Cgmp-Dependent Protein Kinase Pkg From Plasmodium Falciparum - Apo Form
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2015-11-04 Classification: TRANSFERASE Ligands: EDO, SO4, GOL |
 |
Crystal Structure Of The Cgmp-Dependent Protein Kinase Pkg From Plasmodium Vivax - Apo Form
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2015-11-04 Classification: TRANSFERASE |
 |
Co-Crystal Structure Of The N-Termial Cgmp Binding Domain Of Plasmodium Falciparum Pkg With Cgmp
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2015-11-04 Classification: TRANSFERASE Ligands: PCG |
 |
Crystal Structure Of The Cgmp-Dependent Protein Kinase Pkg From Plasmodium Vivax - Amppnp Bound
Organism: Plasmodium vivax (strain salvador i)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-10-14 Classification: TRANSFERASE Ligands: ANP, UNX, CL, NA |
 |
Organism: Bacillus sp. ps3
Method: X-RAY DIFFRACTION Resolution:3.90 Å Release Date: 2015-08-26 Classification: HYDROLASE Ligands: SO4, ADP |
 |
Organism: Zygosaccharomyces rouxii cbs 732
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2015-08-19 Classification: REPLICATION REGULATOR Ligands: GOL |
 |
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-08-19 Classification: REPLICATION REGULATOR Ligands: SO4, GOL |
 |
Crystal Structure Of Human Lysophosphatidic Acid Receptor 1 In Complex With Ono9780307
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2015-06-03 Classification: TRANSPORT PROTEIN/inhibitor Ligands: ON7, 1WV |
 |
Crystal Structure Of Human Lysophosphatidic Acid Receptor 1 In Complex With Ono-9910539
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2015-06-03 Classification: TRANSPORT PROTEIN/inhibitor Ligands: ON9, 1WV |
 |
Crystal Structure Of Human Lysophosphatidic Acid Receptor 1 In Complex With Ono-3080573
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2015-06-03 Classification: TRANSPORT PROTEIN/inhibitor Ligands: ON3, 1WV |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-08-20 Classification: REPLICATION REGULATOR Ligands: SO4, EDO |
 |
Crystal Structure Of The Dna-Binding Domain Of Cole2-P9 Rep In Complex With The Replication Origin
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2013-07-31 Classification: DNA BINDING PROTEIN/DNA Ligands: SO4 |
 |
Organism: Micrococcus luteus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2011-04-13 Classification: HYDROLASE |
 |
Crystal Structure Of Mglu In Its L-Glutamate Binding Form In The Presence Of 4.3M Nacl
Organism: Micrococcus luteus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2011-04-13 Classification: HYDROLASE Ligands: GLU |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2011-04-13 Classification: HYDROLASE |

