Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Lacticaseibacillus rhamnosus
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE |
 |
Organism: Lacticaseibacillus rhamnosus
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE |
 |
Organism: Lacticaseibacillus rhamnosus
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE Ligands: PLP |
 |
Organism: Lacticaseibacillus rhamnosus
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE |
 |
Organism: Corynebacterium glutamicum
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSPORT PROTEIN Ligands: PGT, BET, NA |
 |
Crystal Structure Of The Helicobacter Pylori Copper Resistance Determinant Crda In The Apo Form
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: METAL BINDING PROTEIN Ligands: SO4 |
 |
Crystal Structure Of The Helicobacter Pylori Copper Resistance Determinant Crda In Complex With Silver Ions In Space Group P21
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: METAL BINDING PROTEIN Ligands: AG, SO4 |
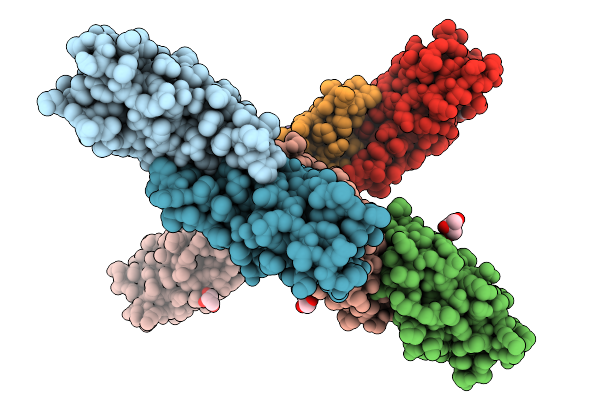 |
Crystal Structure Of The Helicobacter Pylori Copper Resistance Determinant Crda In Complex With Silver Ions In Space Group P1
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: METAL BINDING PROTEIN Ligands: AG, GOL |
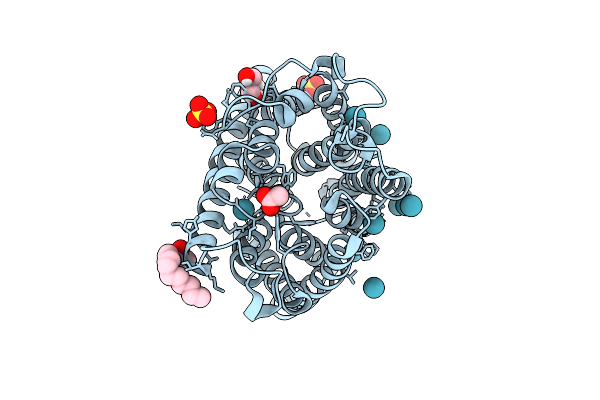 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: LDA, ACY, GOL, XE, SO4, NH2 |
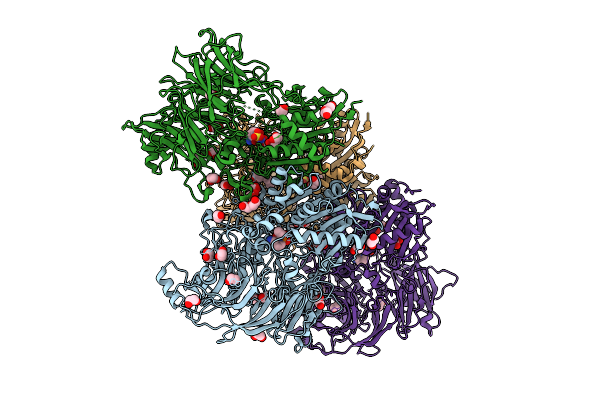 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: A1INE, EDO, PEG, PG5, PGE |
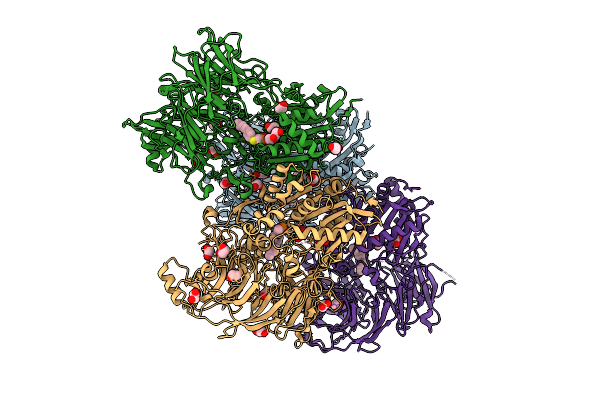 |
Crystal Structure Of Dpp9 In Complex With N-Phosphono-(S)-3-Aminopiperidine-2-One-Based Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: A1INH, EDO, CL |
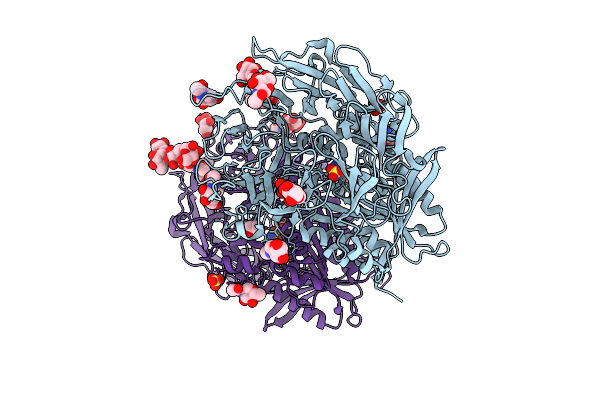 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: NAG, A1INF, SO4, EDO |
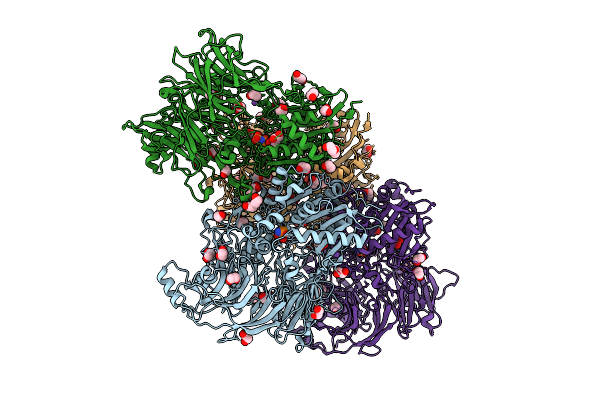 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: A1INF, PEG, EDO, CL, NA, PGE |
 |
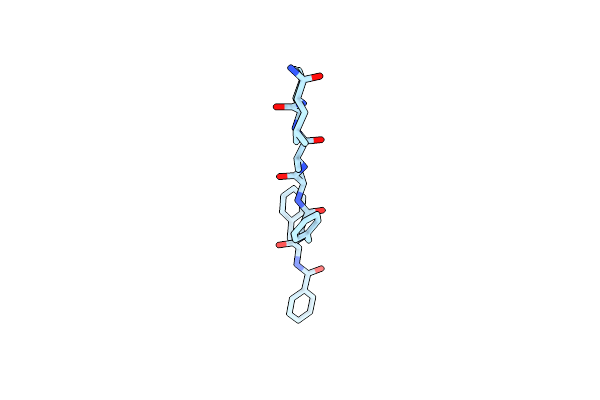
Set Domain Of Histone-Lysine N-Methyltransferase Nsd2 In Complex With Selective Inhibitor
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2025-04-30 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: ZN, A1A0M |
 |
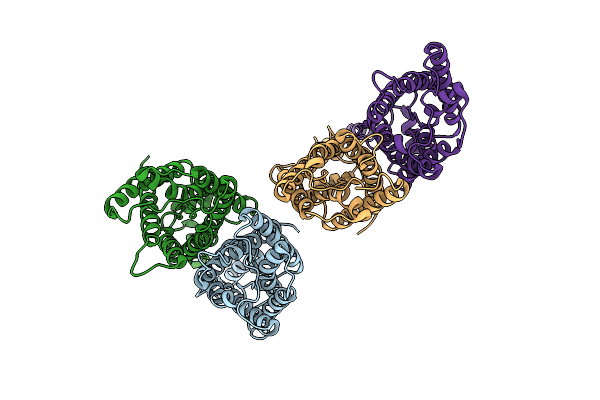
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2025-04-09 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON CRYSTALLOGRAPHY Resolution:2.60 Å Release Date: 2025-02-05 Classification: MEMBRANE PROTEIN |
 |
Organism: Micromonospora pattaloongensis
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2025-01-15 Classification: HYDROLASE Ligands: EDO, GOL |
 |
Organism: Micromonospora pattaloongensis
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2025-01-15 Classification: HYDROLASE |
 |
Organism: Kutzneria buriramensis
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2025-01-15 Classification: HYDROLASE |
 |
Organism: Kutzneria buriramensis
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2025-01-15 Classification: HYDROLASE |