Search Count: 17
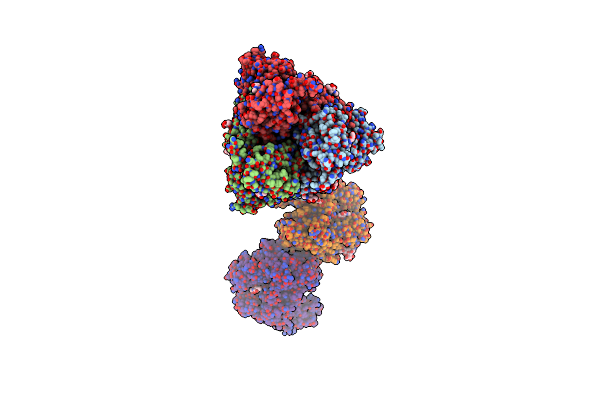 |
Cryoem Structure Of The Spike Protein Of Human Cov 229E In Complex With Receptor Hapn (Composite Map)
Organism: Human coronavirus 229e, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: VIRAL PROTEIN Ligands: NAG |
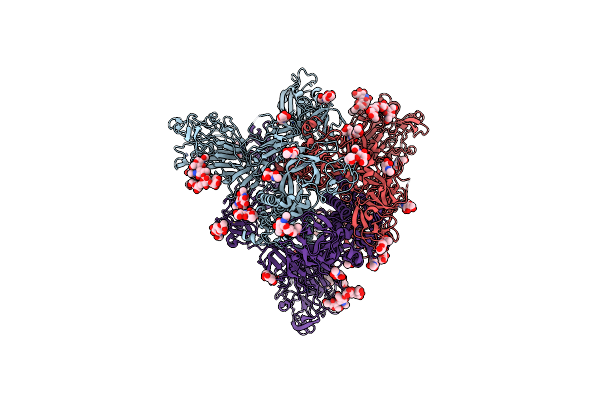 |
Organism: Human betacoronavirus 2c emc/2012
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: VIRAL PROTEIN Ligands: NAG |
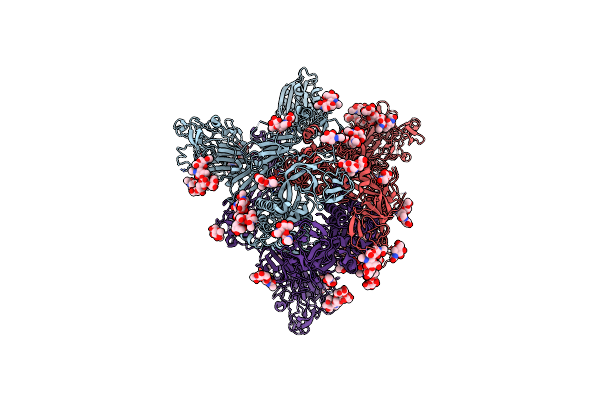 |
Organism: Human betacoronavirus 2c emc/2012
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: VIRAL PROTEIN Ligands: NAG |
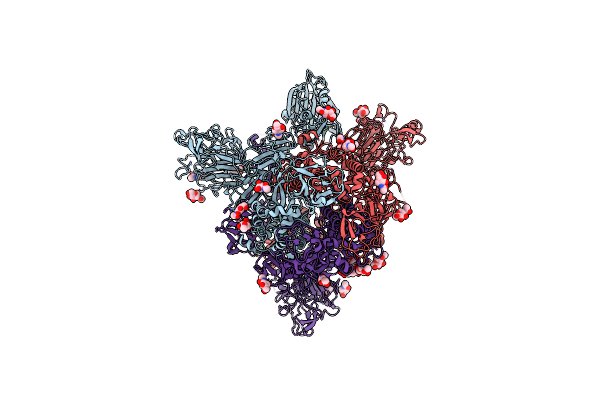 |
Organism: Human betacoronavirus 2c emc/2012
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Human betacoronavirus 2c emc/2012
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Human betacoronavirus 2c emc/2012
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Human betacoronavirus 2c emc/2012
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Human betacoronavirus 2c emc/2012
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: VIRAL PROTEIN Ligands: NAG |
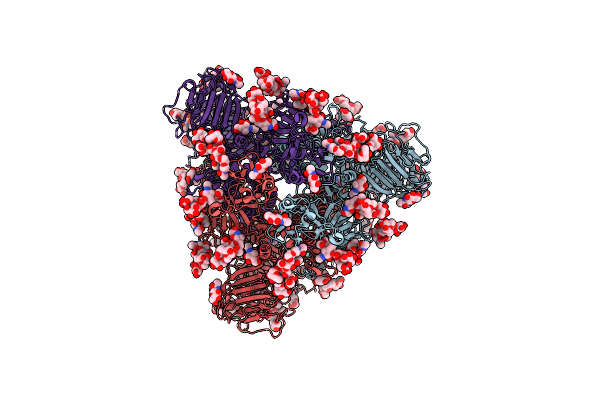 |
Cryo-Em Map Of Pedv (Pintung 52) S Protein With All Three Protomers In The D0-Down Conformation Determined In Situ On Intact Viral Particles.
Organism: Porcine epidemic diarrhea virus
Method: ELECTRON MICROSCOPY Release Date: 2022-08-03 Classification: VIRAL PROTEIN Ligands: NAG |
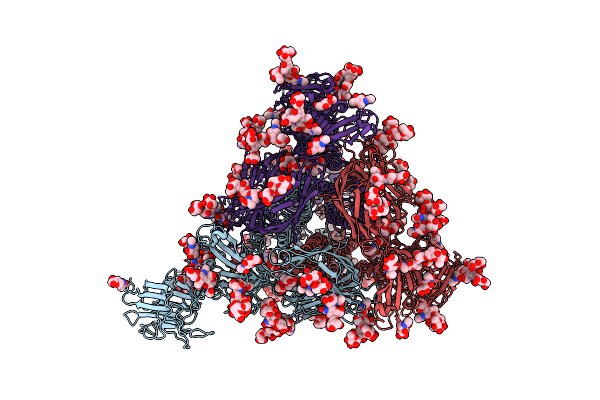 |
Cryo-Em Map Of Pedv S Protein With One Protomer In The D0-Up Conformation While The Other Two In The D0-Down Conformation
Organism: Porcine epidemic diarrhea virus
Method: ELECTRON MICROSCOPY Release Date: 2022-08-03 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Porcine epidemic diarrhea virus
Method: ELECTRON MICROSCOPY Release Date: 2022-08-03 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Cryo-Em Map Of Ipec-J2 Cell-Derived Pedv Pt52 S Protein One D0-Down And Two D0-Up
Organism: Porcine epidemic diarrhea virus
Method: ELECTRON MICROSCOPY Release Date: 2022-08-03 Classification: VIRAL PROTEIN Ligands: NAG |
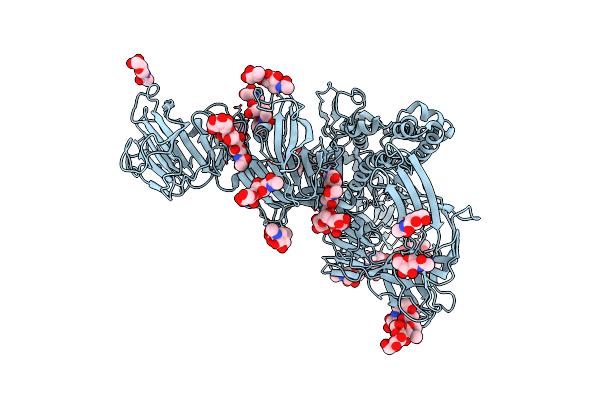 |
Symmetry-Expanded And Locally Refined Protomer Structure Of Ipec-J2 Cell-Derived Pedv Pt52 S With A Ctd-Close Conformation
Organism: Porcine epidemic diarrhea virus
Method: ELECTRON MICROSCOPY Release Date: 2022-08-03 Classification: VIRAL PROTEIN Ligands: NAG |
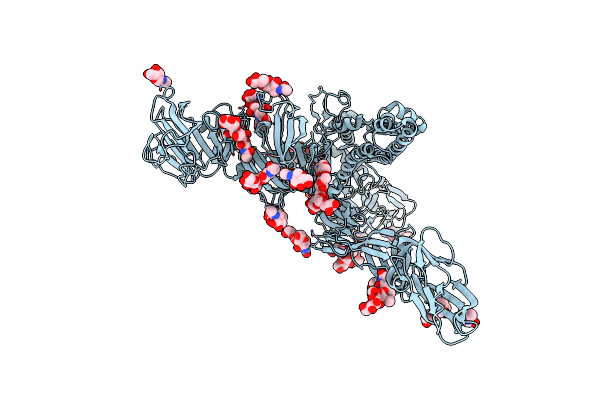 |
Symmetry-Expanded And Locally Refined Protomer Structure Of Ipec-J2 Cell-Derived Pedv Pt52 S With A Ctd-Open Conformation
Organism: Porcine epidemic diarrhea virus
Method: ELECTRON MICROSCOPY Resolution:3.30 Å Release Date: 2022-08-03 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Cryo-Em Structure Of Spike Protein Of Feline Infectious Peritonitis Virus Strain Uu4
Organism: Feline infectious peritonitis virus
Method: ELECTRON MICROSCOPY Release Date: 2020-01-15 Classification: VIRAL PROTEIN Ligands: NAG, MAN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2009-05-12 Classification: CELL CYCLE Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2008-11-11 Classification: HYDROLASE |

