Search Count: 9
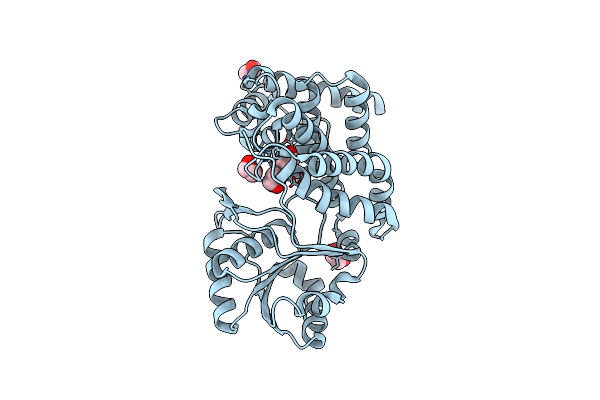 |
Crystal Structure Of Ethylene Glycol-Bound Glycerol Dehydrogenase From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2024-05-29 Classification: OXIDOREDUCTASE Ligands: ZN, EDO, GOL |
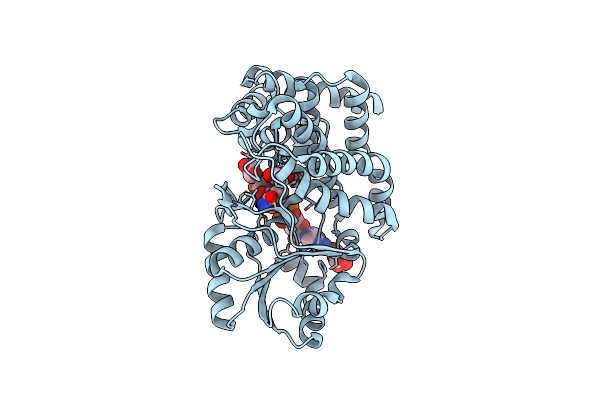 |
Crystal Structure Of Ethylene Glycol-Bound Glycerol Dehydrogenase From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-05-29 Classification: OXIDOREDUCTASE Ligands: ZN, NAD, EDO, TLA |
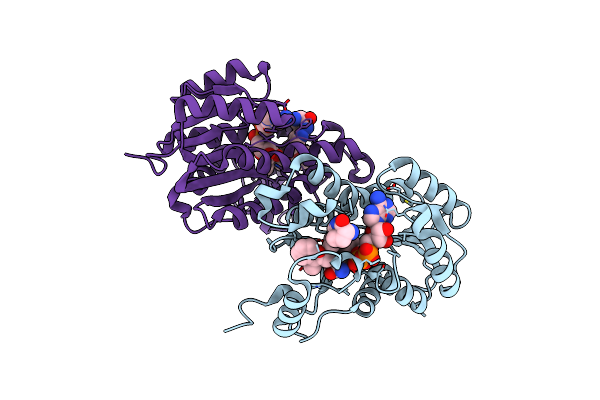 |
Crystal Structure Of E. Coli Fabi In Complex With Nad And (R,E)-3-(7-Amino-8-Oxo-6,7,8,9-Tetrahydro-5H-Pyrido[2,3-B]Azepin-3-Yl)-N-Methyl-N-((3-Methylbenzofuran-2-Yl)Methyl)Acrylamide
Organism: Escherichia coli str. k-12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-02-15 Classification: ANTIBIOTIC Ligands: NAD, NQF |
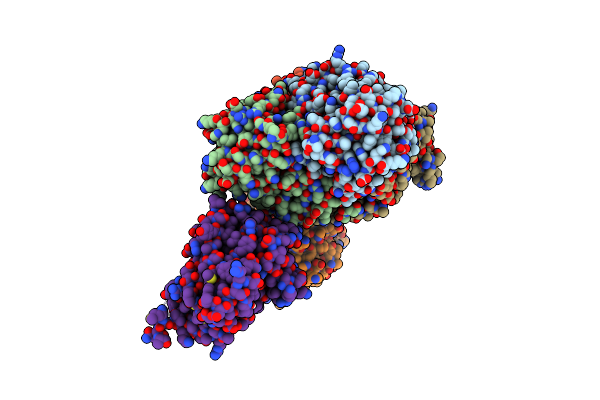 |
Crystal Structure Of Acinetobacter Baumannii Fabi In Complex With Nad And (R,E)-3-(7-Amino-8-Oxo-6,7,8,9-Tetrahydro-5H-Pyrido[2,3-B]Azepin-3-Yl)-N-Methyl-N-((3-Methylbenzofuran-2-Yl)Methyl)Acrylamide
Organism: Acinetobacter baumannii atcc 19606 = cip 70.34 = jcm 6841
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2023-02-15 Classification: ANTIBIOTIC Ligands: NQF, NAD |
 |
Crystal Structure Of Acinetobacter Baumannii Fabi In Complex With Nad And Fabimycin ((S,E)-3-(7-Amino-8-Oxo-6,7,8,9-Tetrahydro-5H-Pyrido[2,3-B]Azepin-3-Yl)-N-Methyl-N-((3-Methylbenzofuran-2-Yl)Methyl)Acrylamide)
Organism: Acinetobacter baumannii atcc 19606 = cip 70.34 = jcm 6841
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2023-02-15 Classification: ANTIBIOTIC Ligands: NAD, NUC |
 |
Crystal Structure Of E. Coli Fabi In Complex With Nad And Fabimycin ((S,E)-3-(7-Amino-8-Oxo-6,7,8,9-Tetrahydro-5H-Pyrido[2,3-B]Azepin-3-Yl)-N-Methyl-N-((3-Methylbenzofuran-2-Yl)Methyl)Acrylamide)
Organism: Escherichia coli str. k-12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2022-12-14 Classification: ANTIBIOTIC Ligands: NAD, NUC |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-06-05 Classification: HYDROLASE Ligands: IPA |
 |
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2018-03-14 Classification: TOXIN Ligands: ZN |
 |
Crystal Structure Of The Zinc-Free Catalytic Domain Of Botulinum Neurotoxin X
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-03-14 Classification: TOXIN Ligands: PEG |

