Search Count: 30
 |
Computational Design Of Stable Mammalian Serum Albumins For Bacterial Expression
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-05-10 Classification: TRANSPORT PROTEIN Ligands: MYR, RWF, DAO |
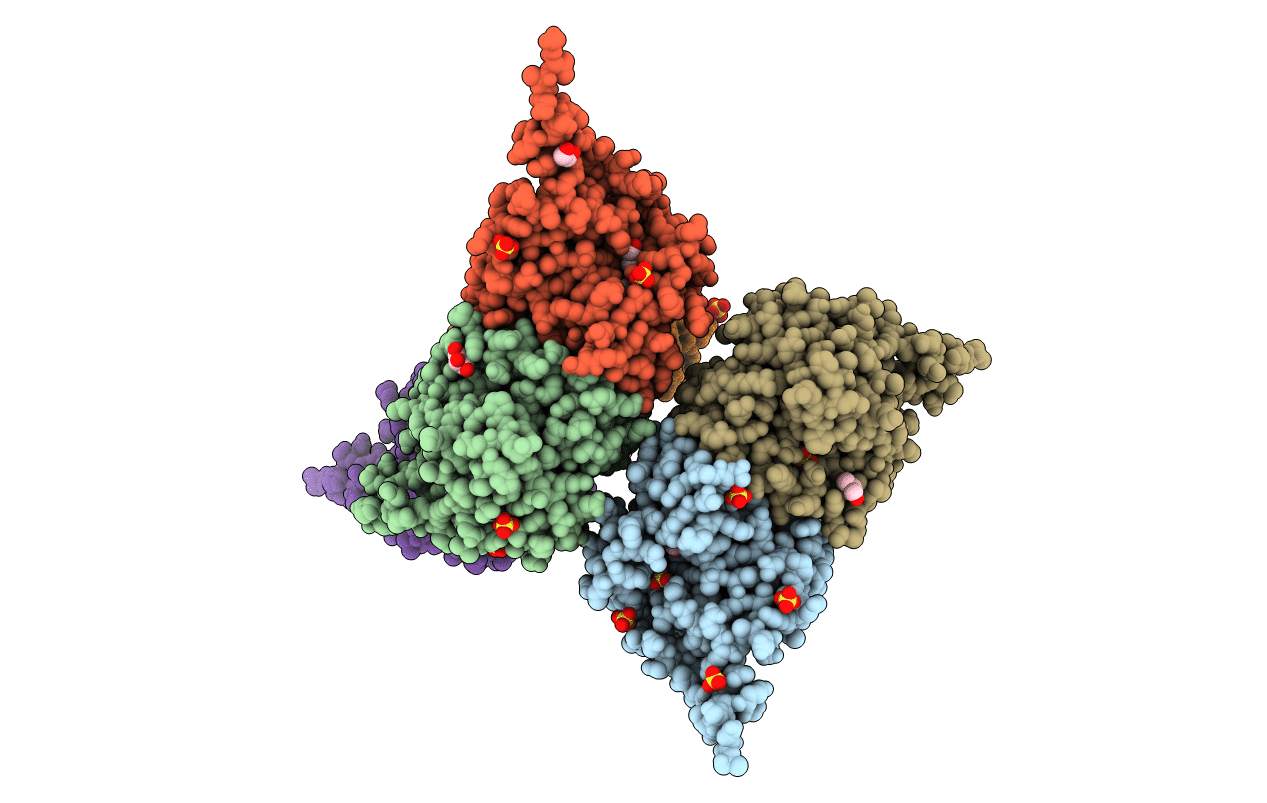 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-03-31 Classification: TRANSFERASE Ligands: 8LK, SO4, GOL |
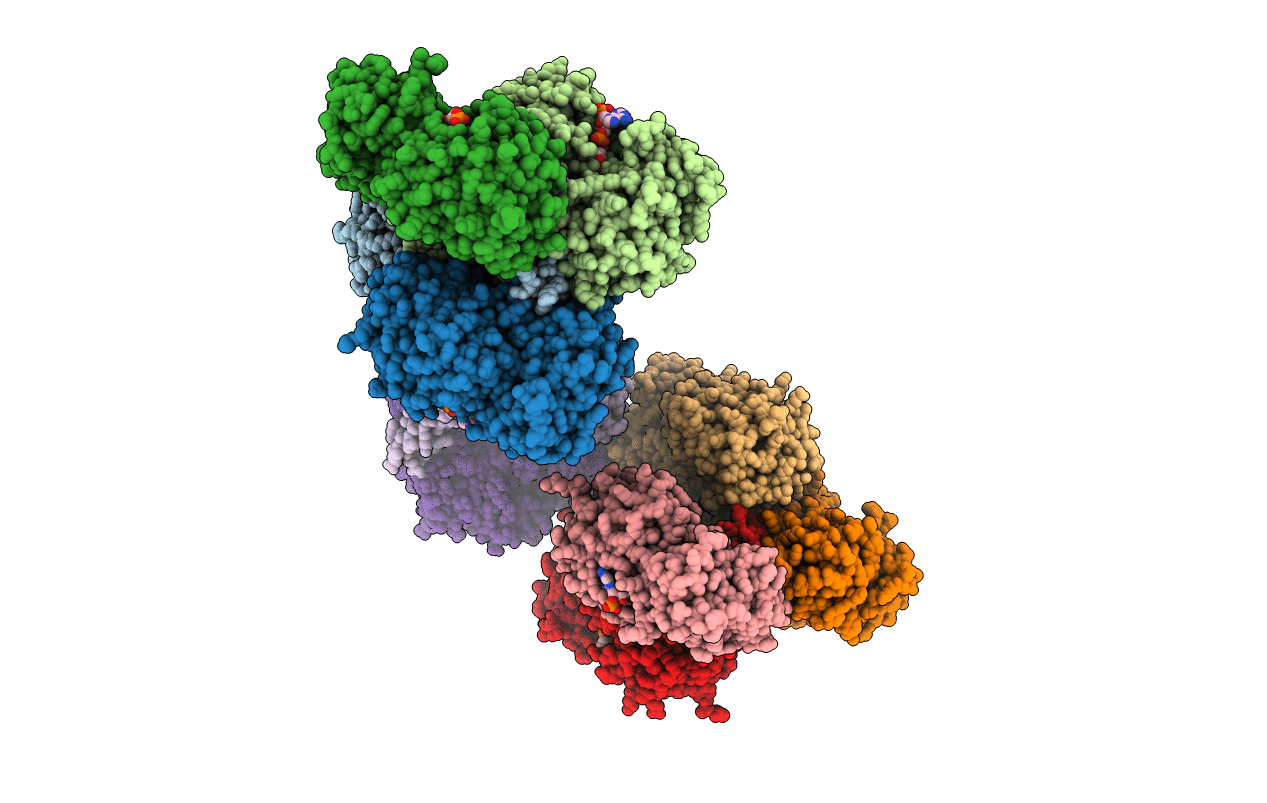 |
Organism: Rhodopseudomonas palustris bisb18
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2018-11-28 Classification: OXIDOREDUCTASE Ligands: NAP, K |
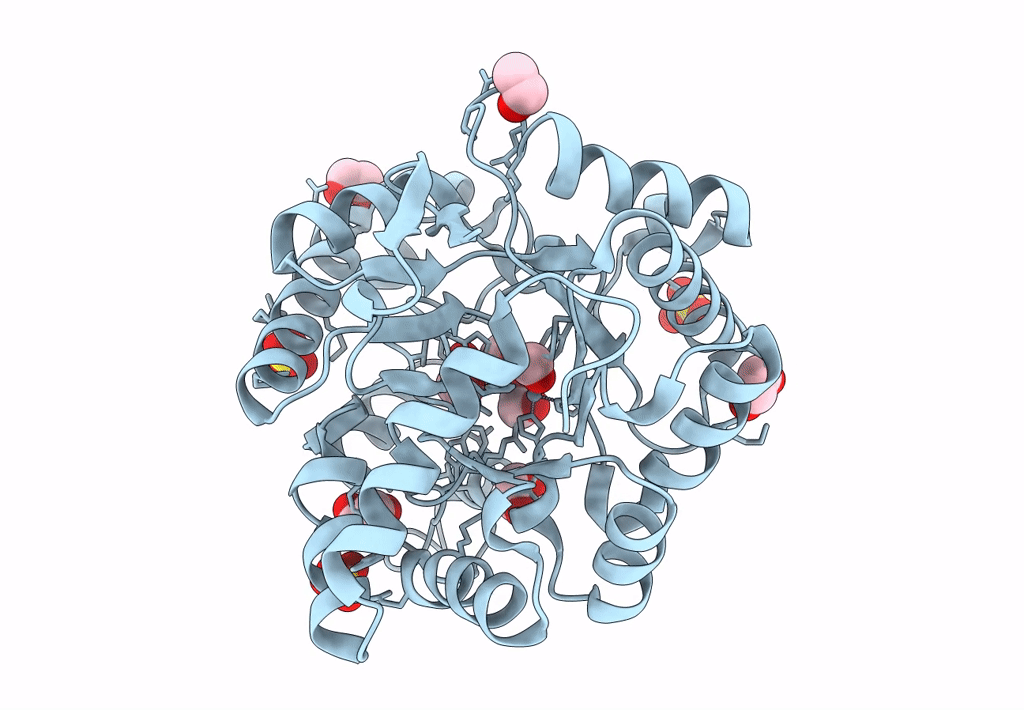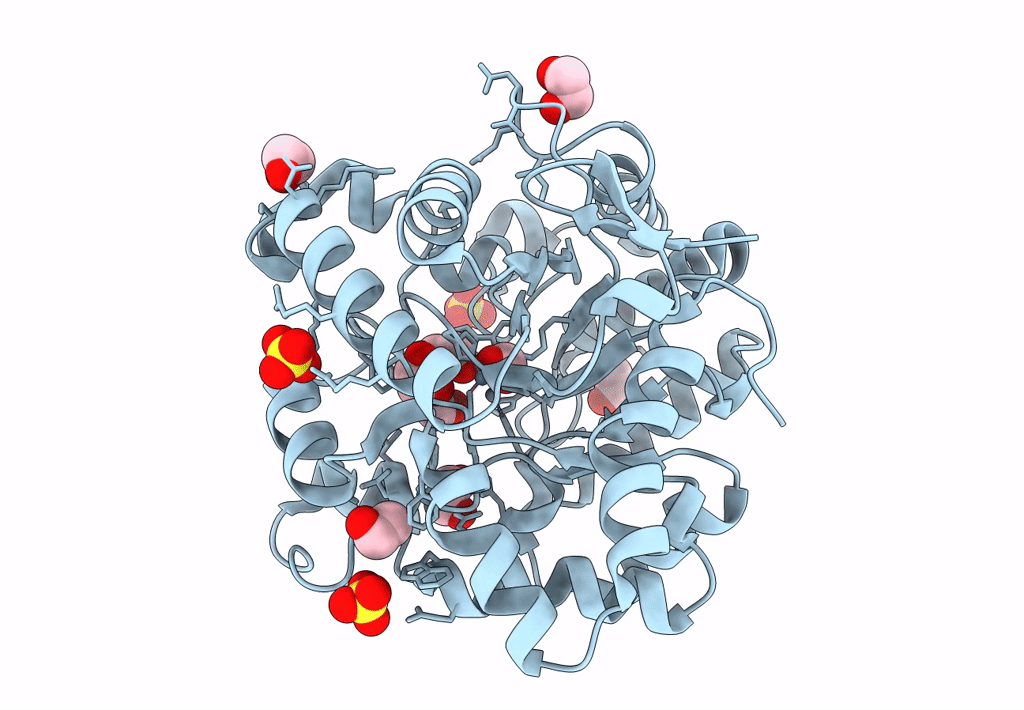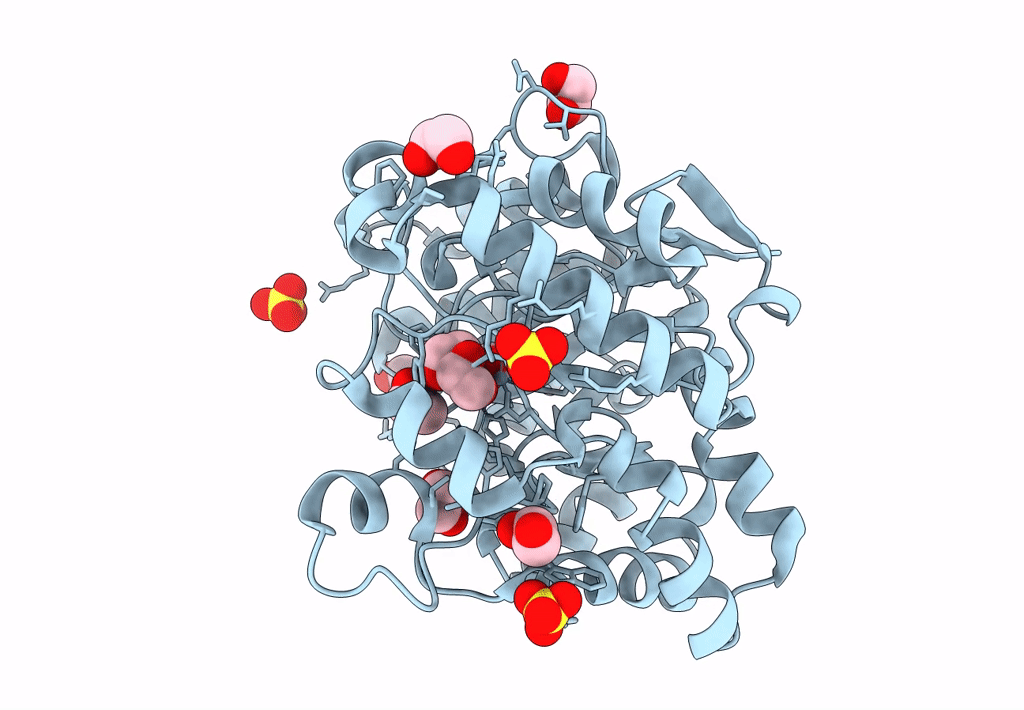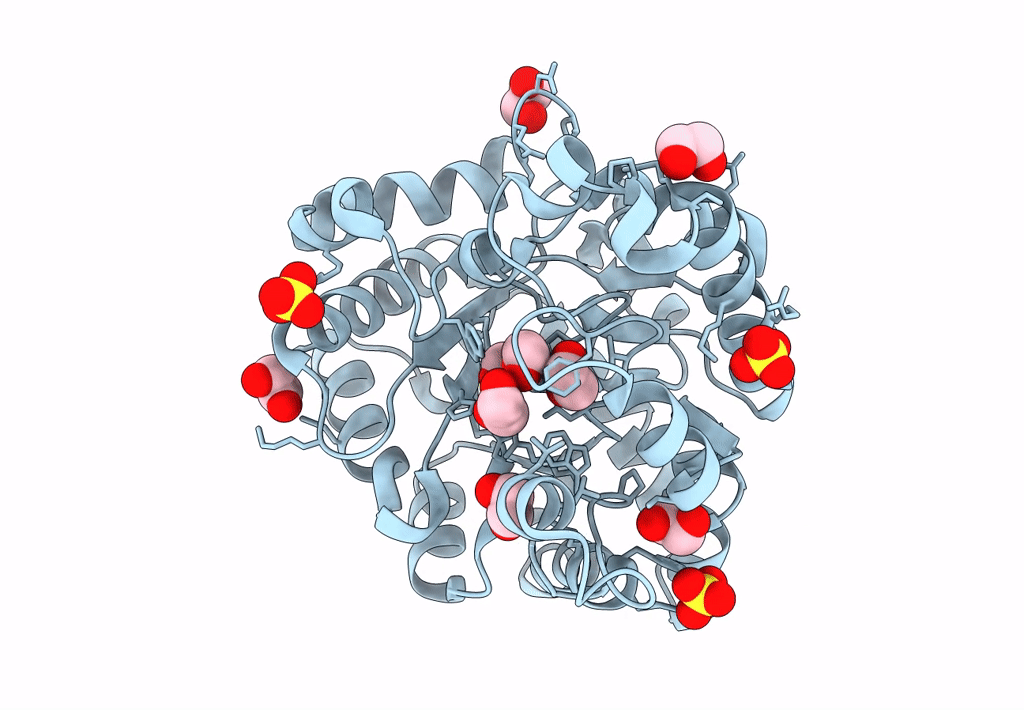 |
Repertoires Of Functionally Diverse Enzymes Through Computational Design At Epistatic Active-Site Positions
Organism: Brevundimonas diminuta
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2018-10-24 Classification: HYDROLASE Ligands: FMT, ZN, EDO, SO4 |
 |
Repertoires Of Functionally Diverse Enzymes Through Computational Design At Epistatic Active-Site Positions
Organism: Brevundimonas diminuta
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-10-24 Classification: HYDROLASE Ligands: FMT, ZN, TRS |
 |
Repertoires Of Functionally Diverse Enzymes Through Computational Design At Epistatic Active-Site Positions
Organism: Brevundimonas diminuta
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2018-10-24 Classification: HYDROLASE Ligands: FMT, ZN, EDO, CAC |
 |
Highly Active Enzymes By Automated Modular Backbone Assembly And Sequence Design
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2018-07-25 Classification: Artificial enzyme |
 |
Highly Active Enzymes By Automated Modular Backbone Assembly And Sequence Design
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2018-07-25 Classification: Artificial enzyme |
 |
X-Ray Structure Of The De-Novo Design Amidase At The Resolution 1.8A, Northeast Structural Genomics Consortium (Nesg) Target Or398
Organism: Artificial gene
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2013-08-07 Classification: DE NOVO PROTEIN Ligands: CL, PEG |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2012-06-27 Classification: LYASE Ligands: SO4, NA |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-06-06 Classification: LYASE Ligands: PO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-06-06 Classification: LYASE/LYASE INHIBITOR Ligands: 0CU, PO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2012-06-06 Classification: LYASE Ligands: SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-06-06 Classification: LYASE Ligands: PO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-06-06 Classification: LYASE/LYASE INHIBITOR Ligands: 0CU, PO4, NA |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2012-06-06 Classification: LYASE/LYASE INHIBITOR Ligands: 0CT, PO4 |
 |
Optimization Of The In Silico Designed Kemp Eliminase Ke70 By Computational Design And Directed Evolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2011-02-09 Classification: LYASE |
 |
Optimization Of The In Silico Designed Kemp Eliminase Ke70 By Computational Design And Directed Evolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2011-02-09 Classification: LYASE |
 |
In Silico Designed Of An Improved Kemp Eliminase Ke70 Mutant By Computational Design And Directed Evolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2011-02-09 Classification: LYASE |
 |
Optimization Of The In Silico Designed Kemp Eliminase Ke70 By Computational Design And Directed Evolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2011-02-09 Classification: LYASE |

