Search Count: 14
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2020-03-04 Classification: APOPTOSIS/INHIBITOR Ligands: Q0D |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2020-03-04 Classification: APOPTOSIS/INHIBITOR Ligands: Q0G |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2020-03-04 Classification: APOPTOSIS/INHIBITOR Ligands: Q0A, EDO, ACT, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2020-03-04 Classification: APOPTOSIS/INHIBITOR Ligands: Q01, BNL |
 |
The Crystal Structure Of Anti-Apoptotic Mcl-1 Protein In Complex With 2, 5-Substituted Benzoic Acid Inhibitor 21
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2020-03-04 Classification: APOPTOSIS/INHIBITOR Ligands: PZY |
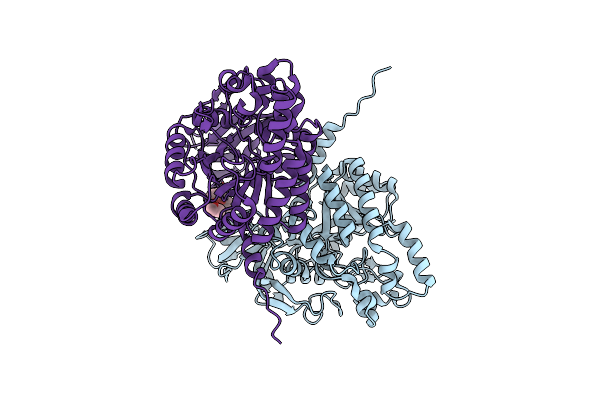 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2017-03-22 Classification: SIGNALING PROTEIN Ligands: 1PG |
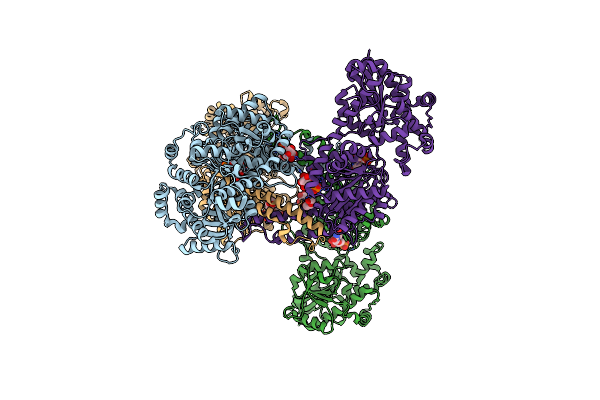 |
Crystal Structure Of Yeast Glycogen Synthase In Complex With Uridine-5'-Monophosphate
Organism: Saccharomyces cerevisiae fosterso
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2013-12-11 Classification: TRANSFERASE Ligands: U5P, G6P, PEG |
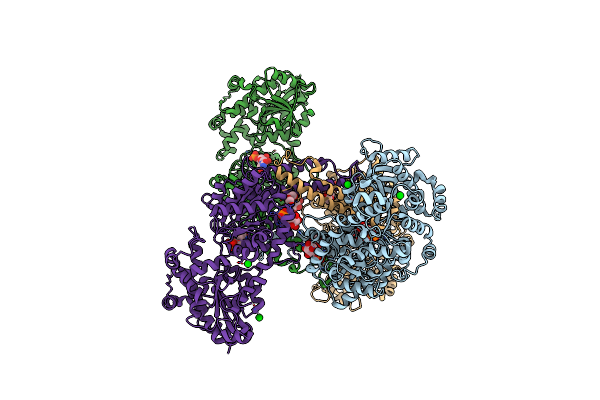 |
Glucose1,2Cyclic Phosphate Bound Activated State Of Yeast Glycogen Synthase
Organism: Saccharomyces cerevisiae fosterso
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2013-12-11 Classification: TRANSFERASE Ligands: U5P, G6P, PEG, BA, 1S3 |
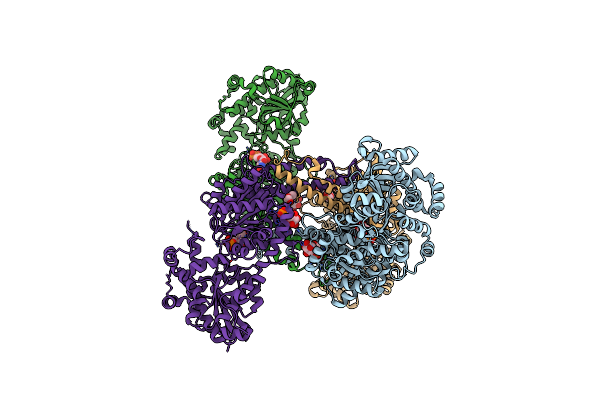 |
Crystal Structure Of Yeast Glycogen Synthase E169Q Mutant In Complex With Glucose And Udp
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2013-12-11 Classification: TRANSFERASE Ligands: UDP, G6P, PEG, GLC, BGC |
 |
Crystal Structure Of Human Aldh2 Modified With The Beta-Elimination Product Of Aldi-3; 1-(4-Ethylbenzene)Prop-2-En-1-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-11-02 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2011-11-02 Classification: OXIDOREDUCTASE Ligands: K, ACT |
 |
Crystal Structure Of Human Aldh3A1 Modified With The Beta-Elimination Product Of Aldi-1; 1-Phenyl- 2-Propen-1-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2011-11-02 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: K, ACT, I1E |
 |
Solution Structure Of The Internal Loop Of Human U65 H/Aca Snorna 3' Hairpin
|
 |
Structural Characterization Of Nop10P Using Nuclear Magnetic Resonance Spectroscopy
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2005-12-06 Classification: BIOSYNTHETIC PROTEIN |

