Search Count: 34
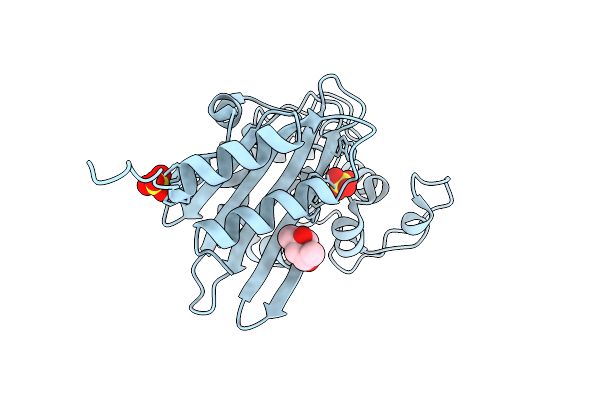 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: HYDROLASE Ligands: SO4, MPD |
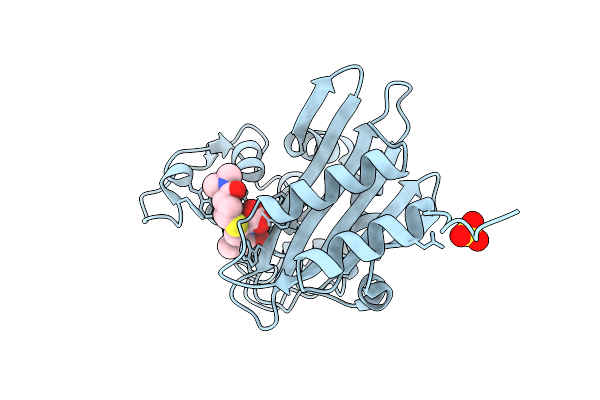 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: HYDROLASE Ligands: Y33, SO4 |
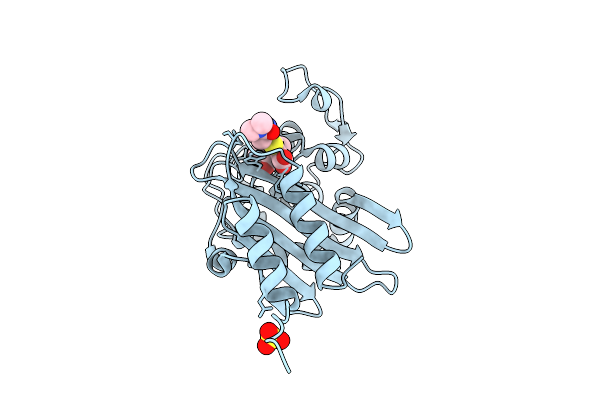 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: HYDROLASE Ligands: Y33, SO4 |
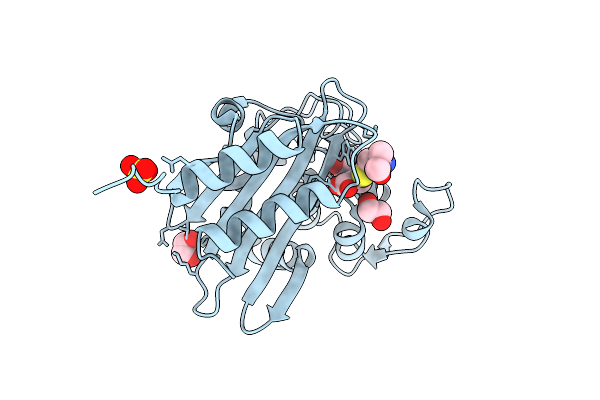 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: HYDROLASE Ligands: Y33, SO4, EDO |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: HYDROLASE Ligands: MER, SO4 |
 |
Organism: Lama glama, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: PROTON TRANSPORT |
 |
Organism: Lama glama, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: PROTON TRANSPORT |
 |
Organism: Lama glama, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Resolution:3.70 Å Release Date: 2025-02-05 Classification: PROTON TRANSPORT |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-12-11 Classification: HYDROLASE/INHIBITOR |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-12-11 Classification: HYDROLASE/INHIBITOR Ligands: Y33, GOL |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-12-11 Classification: HYDROLASE/INHIBITOR Ligands: Y33 |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2024-12-11 Classification: HYDROLASE/INHIBITOR Ligands: Y33, ACT |
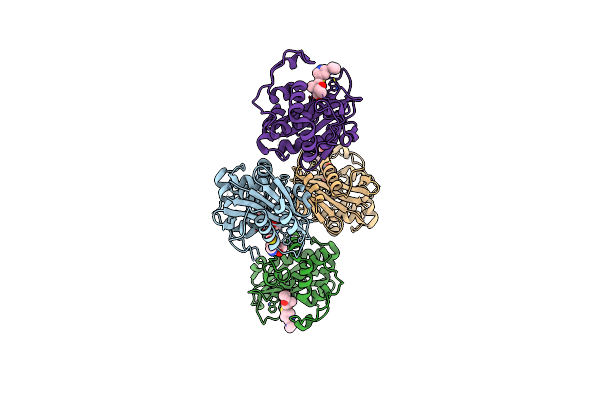 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2024-12-11 Classification: HYDROLASE/INHIBITOR Ligands: Y33, CO2 |
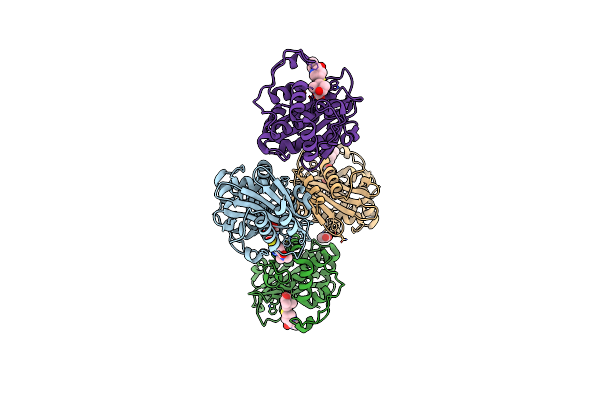 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-12-11 Classification: HYDROLASE/INHIBITOR Ligands: Y33, CO2, ACT |
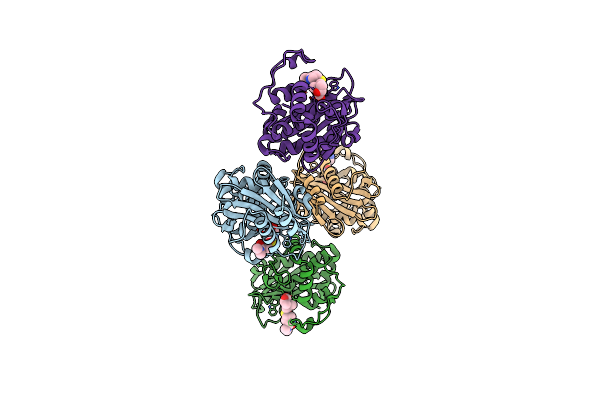 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2024-12-11 Classification: HYDROLASE/INHIBITOR Ligands: Y33 |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2021-12-29 Classification: MOTOR PROTEIN |
 |
Organism: Saccharomyces cerevisiae s288c, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-12-22 Classification: MOTOR PROTEIN |
 |
Organism: Saccharomyces cerevisiae s288c, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-12-22 Classification: MOTOR PROTEIN |
 |
Organism: Saccharomyces cerevisiae s288c, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-12-22 Classification: MOTOR PROTEIN |
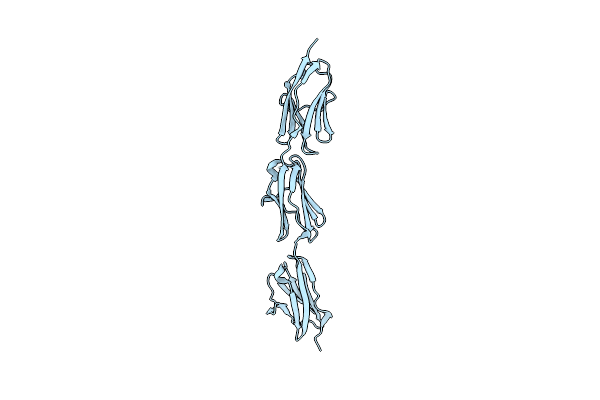 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2016-09-14 Classification: STRUCTURAL PROTEIN |

