Search Count: 26
 |
Organism: Fusobacterium nucleatum subsp. nucleatum (strain atcc 25586 / cip 101130 / jcm 8532 / lmg 13131)
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2021-02-17 Classification: LYASE Ligands: MPD, NO3 |
 |
Organism: Porphyromonas gingivalis (strain atcc 33277 / dsm 20709 / cip 103683 / jcm 12257 / nctc 11834 / 2561)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-04-24 Classification: TRANSFERASE |
 |
Crystal Structure Of Porphyromonas Gingivalis Phosphotransacetylase In Complex With Acetyl-Coa
Organism: Porphyromonas gingivalis (strain atcc 33277 / dsm 20709 / cip 103683 / jcm 12257 / nctc 11834 / 2561)
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2019-04-24 Classification: TRANSFERASE Ligands: ACO |
 |
Organism: Porphyromonas gingivalis (strain atcc 33277 / dsm 20709 / cip 103683 / jcm 12257 / nctc 11834 / 2561)
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2019-04-24 Classification: TRANSFERASE Ligands: MPD, SO4 |
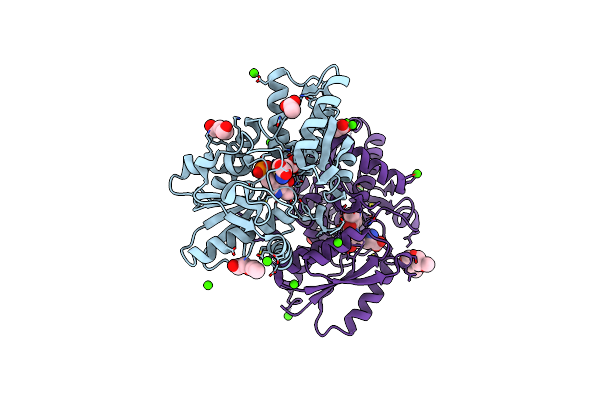 |
Crystal Structure Of A Hydrogen Sulfide-Producing Enzyme (Fn1220) From Fusobacterium Nucleatum In Complex With L-Lanthionine-Plp Schiff Base
Organism: Fusobacterium nucleatum subsp. nucleatum strain atcc 25586
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2018-04-11 Classification: TRANSFERASE Ligands: 85F, PLP, ACT, CA, PEG, PG0, PG5 |
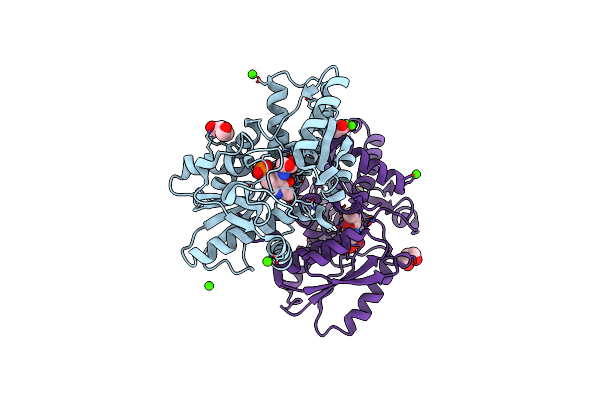 |
Crystal Structure Of A Hydrogen Sulfide-Producing Enzyme (Fn1220) From Fusobacterium Nucleatum In Complex With L-Serine-Plp Schiff Base
Organism: Fusobacterium nucleatum subsp. nucleatum strain atcc 25586
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2018-04-11 Classification: TRANSFERASE Ligands: PLP, SER, ACT, CA, PEG |
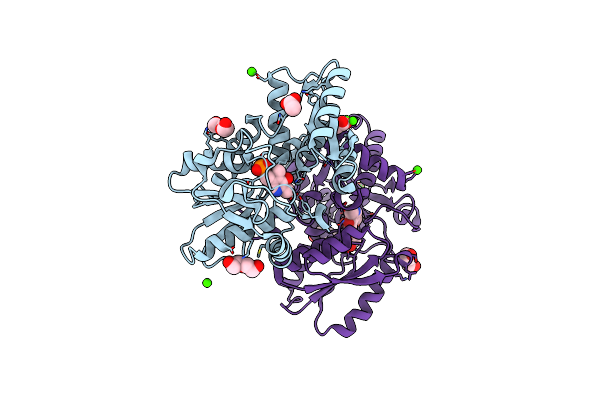 |
Crystal Structure Of A Hydrogen Sulfide-Producing Enzyme (Fn1220) From Fusobacterium Nucleatum
Organism: Fusobacterium nucleatum subsp. nucleatum strain atcc 25586
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2018-04-11 Classification: TRANSFERASE Ligands: PLP, ACT, CA, PEG |
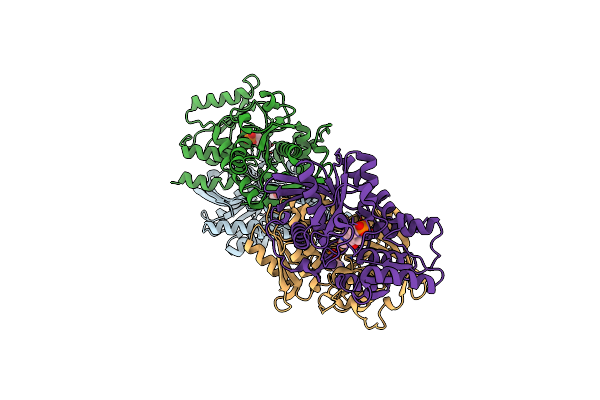 |
Crystal Structure Of Hydrogen Sulfide-Producing Enzyme (Fn1055) From Fusobacterium Nucleatum: Lysine-Dimethylated Form
Organism: Fusobacterium nucleatum subsp. nucleatum (strain atcc 25586 / cip 101130 / jcm 8532 / lmg 13131)
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2018-02-14 Classification: TRANSFERASE Ligands: PLP, CL |
 |
Crystal Structure Of Hydrogen Sulfide-Producing Enzyme (Fn1055) From Fusobacterium Nucleatum
Organism: Fusobacterium nucleatum subsp. nucleatum atcc 25586
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2017-04-26 Classification: TRANSFERASE Ligands: PLP, CL |
 |
Crystal Structure Of Hydrogen Sulfide-Producing Enzyme (Fn1055) D232N Mutant In Complexed With Alpha-Aminoacrylate Intermediate: Lysine-Dimethylated Form
Organism: Fusobacterium nucleatum subsp. nucleatum strain atcc 25586
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2017-04-26 Classification: TRANSFERASE Ligands: PLP, 0JO, PEG |
 |
Organism: Solanum lycopersicum
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2014-08-13 Classification: TRANSFERASE |
 |
Organism: Solanum lycopersicum
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2014-08-13 Classification: TRANSFERASE Ligands: TLA |
 |
Organism: Solanum lycopersicum, Tomato mosaic virus (strain l)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-08-13 Classification: TRANSFERASE Ligands: AGS, MG, CS |
 |
Organism: Solanum lycopersicum, Tomato mosaic virus (strain l)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-08-13 Classification: TRANSFERASE Ligands: AGS, MG, CL |
 |
Crystal Structure Of Betac-S Lyase From Streptococcus Anginosus: Internal Aldimine Form
Organism: Streptococcus anginosus
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2012-06-27 Classification: LYASE Ligands: PLP, SO4, GOL |
 |
Crystal Structure Of Betac-S Lyase From Streptococcus Anginosus In Complex With L-Serine: External Aldimine Form
Organism: Streptococcus anginosus
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2012-06-27 Classification: LYASE Ligands: PLP, PLS, EPE, NA |
 |
Crystal Structure Of Betac-S Lyase From Streptococcus Anginosus In Complex With L-Serine: Alpha-Aminoacrylate Form
Organism: Streptococcus anginosus
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2012-06-27 Classification: LYASE Ligands: 0JO, EPE, NA, ACT |
 |
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2010-04-21 Classification: HYDROLASE Ligands: MES, MPD |
 |
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Resolution:3.02 Å Release Date: 2009-05-26 Classification: UNKNOWN FUNCTION Ligands: GOL, SO4 |
 |
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-05-01 Classification: HYDROLASE Ligands: MES, MPD |

