Search Count: 72
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: TRANSPORT PROTEIN Ligands: 3PE, CLR |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: TRANSPORT PROTEIN Ligands: Y01, 3PE |
 |
Cryo-Em Structure Of Mmcat1 Bound With Frmlv-Rbd In The Apo Inward-Open State
Organism: Mus musculus, Murine leukemia virus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: MEMBRANE PROTEIN Ligands: NAG, Y01 |
 |
Cryo-Em Structure Of Mmcat1 Bound With Frmlv-Rbd In The Arginine-Bound Inward-Occluded State
Organism: Mus musculus, Murine leukemia virus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: MEMBRANE PROTEIN Ligands: NAG, Y01, ARG |
 |
Cryo-Em Structure Of Mmcat1 Bound With Frmlv-Rbd In The Lysine-Bound Inward-Occluded State
Organism: Mus musculus, Murine leukemia virus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: MEMBRANE PROTEIN Ligands: LYS, Y01 |
 |
Cryo-Em Structure Of Mmcat1 Bound With Frmlv-Rbd In The Ornithine-Bound Inward-Occluded State
Organism: Mus musculus, Murine leukemia virus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: MEMBRANE PROTEIN Ligands: Y01, ORN |
 |
Organism: Homo sapiens, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: LIPID TRANSPORT |
 |
Organism: Homo sapiens, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: LIPID TRANSPORT Ligands: LMT |
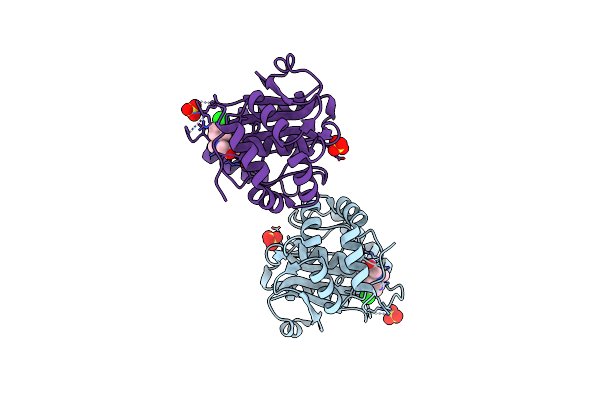 |
Crystal Structure Of Covalent Inhibitor 2-Chloro-N'-(N-(4-Chlorophenyl)-N-Methylglycyl)Acetohydrazide Bound To Ubiquitin C-Terminal Hydrolase-L1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-09-20 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: WEU, SO4 |
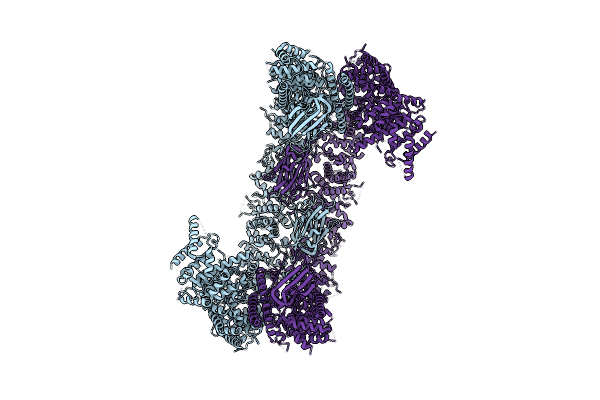 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-08 Classification: APOPTOSIS |
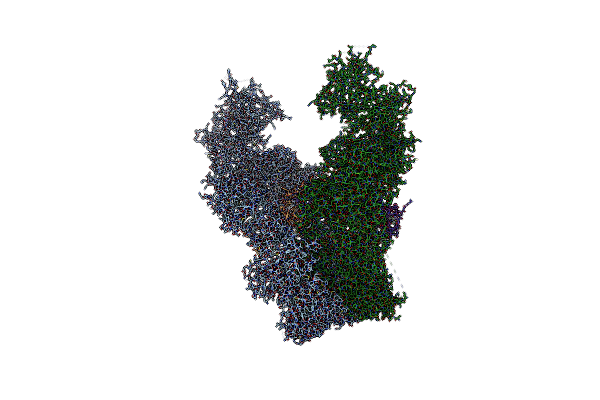 |
Structure Of The Giant Inhibitor Of Apoptosis, Birc6 Bound To The Regulator Smac
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-08 Classification: APOPTOSIS |
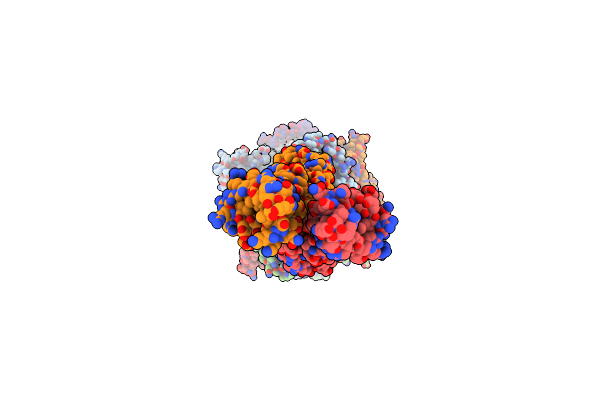 |
Chaetomium Thermophilum Mre11-Rad50-Nbs1 Complex Bound To Atpys (Composite Structure)
Organism: Thermochaetoides thermophila
Method: ELECTRON MICROSCOPY Release Date: 2023-01-11 Classification: HYDROLASE Ligands: MN, MG, AGS |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-01-11 Classification: HYDROLASE Ligands: MN |
 |
Organism: Thermochaetoides thermophila
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: ZN |
 |
Endonuclease State Of The E. Coli Mre11-Rad50 (Sbccd) Head Complex Bound To Adp And Dsdna
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-08-17 Classification: DNA BINDING PROTEIN Ligands: ADP, MG, MN |
 |
Hairpin-Bound State Of The E. Coli Mre11-Rad50 (Sbccd) Head Complex Bound To Adp And A Dna Hairpin
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-08-17 Classification: DNA BINDING PROTEIN Ligands: MN, ADP, MG |
 |
Endonuclease State Of The E. Coli Mre11-Rad50 (Sbccd) Head Complex Bound To Adp And Extended Dsdna
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-08-17 Classification: DNA BINDING PROTEIN Ligands: ADP, MG, MN |
 |
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: ELECTRON MICROSCOPY Release Date: 2019-10-30 Classification: TRANSFERASE Ligands: AGS, MG |
 |
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: ELECTRON MICROSCOPY Release Date: 2019-10-30 Classification: TRANSFERASE Ligands: AGS, MG |
 |
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: ELECTRON MICROSCOPY Release Date: 2019-10-30 Classification: TRANSFERASE Ligands: AGS, MG |

