Search Count: 38
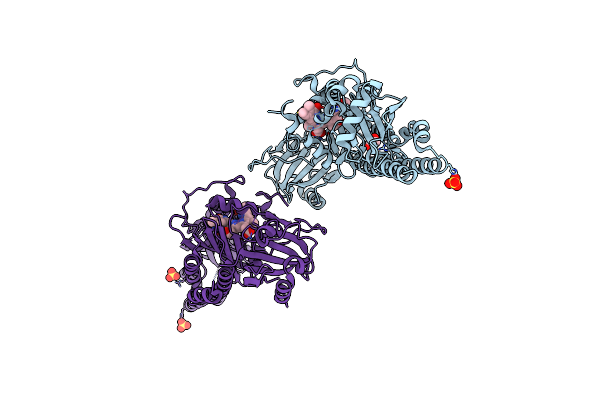 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Pfr State (I0A).
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL, EDO |
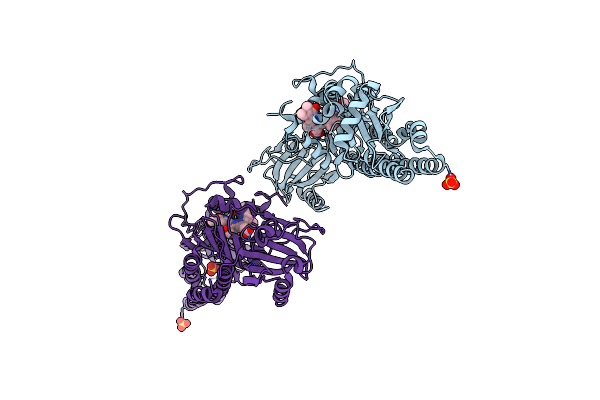 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Pfr State (I0B).
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4 |
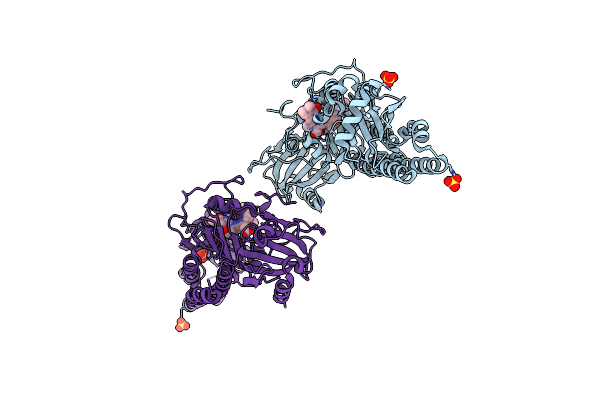 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I1 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4 |
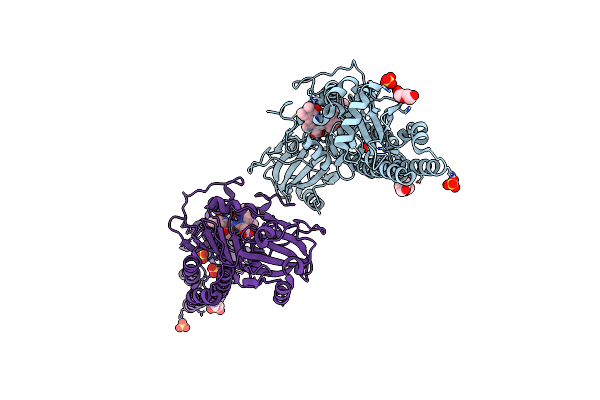 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I2 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, PGE, PEG, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I3 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, GOL, PEG |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I4 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL, PEG |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I5 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I6 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I7 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, PEG |
 |
Time-Resolved Sfx-Xfel Crystal Structure Of Cyp121 Bound With Cyy Reacted With Peracetic Acid For 200 Milliseconds
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: SO4, HEM, YTT, PEO |
 |
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: SO4, HEM, YTT |
 |
Structure Of Xenon-Derivatized Methyl-Coenzyme M Reductase From Methanothermobacter Marburgensis
Organism: Methanothermobacter marburgensis str. marburg
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-04-06 Classification: TRANSFERASE Ligands: F43, MG, ACT, TP7, COM, NA, XE |
 |
Xfel Serial Crystallography Reveals The Room Temperature Structure Of Methyl-Coenzyme M Reductase
Organism: Methanothermobacter marburgensis str. marburg
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-03-02 Classification: TRANSFERASE Ligands: MG, F43, TP7, COM, ACT, EDO |
 |
Room Temperature Xfel Isopenicillin N Synthase Structure In Complex With The Fe(Iv)=O Mimic Vo And Acv.
Organism: Emericella nidulans (strain fgsc a4 / atcc 38163 / cbs 112.46 / nrrl 194 / m139)
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2022-01-12 Classification: OXIDOREDUCTASE Ligands: SO4, ACV, VVO |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-07-21 Classification: ANTIMICROBIAL PROTEIN Ligands: SO4, CL |
 |
Xfel Structure Of Apo Ctx-M-15 After Mixing For 0.7 Sec With Ertapenem Using A Piezoelectric Injector (Polypico)
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2021-07-21 Classification: ANTIMICROBIAL PROTEIN Ligands: SO4 |
 |
Xfel Structure Of The Ertapenem-Derived Ctx-M-15 Acylenzyme After Mixing For 2 Sec Using A Piezoelectric Injector (Polypico)
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2021-07-21 Classification: ANTIMICROBIAL PROTEIN Ligands: TVE, CL, NA |
 |
Room Temperature, Serial X-Ray Structure Of Ctx-M-15 Collected On Fixed Target Chips At Diamond Light Source I24
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-07-21 Classification: ANTIMICROBIAL PROTEIN Ligands: SO4, CL, NA |
 |
Room Temperature, Serial X-Ray Structure Of The Ertapenem-Derived Acylenzyme Of Ctx-M-15 (10 Min Soak) Collected On Fixed Target Chips At Diamond Light Source I24
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-07-21 Classification: ANTIMICROBIAL PROTEIN Ligands: TVE, CL, NA |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2021-07-21 Classification: HYDROLASE Ligands: CL, NA |

