Search Count: 18
 |
Crystal Structure Of Benzoylformate Decarboxylase In Complex With The Inhibitor Mbp
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2009-01-27 Classification: LYASE Ligands: D7K, CA |
 |
Crystal Structure Of Benzoylformate Decarboxylase In Complex With The Pyridyl Inhibitor Paa
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2008-12-09 Classification: LYASE Ligands: MG, 8PA |
 |
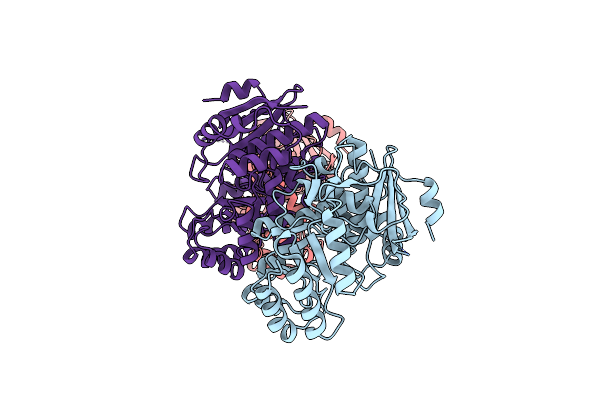
Crystal Structure Of Benzoylformate Decarboxylase In Complex With The Pyridyl Inhibitor 3-Pkb
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2008-12-09 Classification: LYASE Ligands: MG, 8PA |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2008-07-01 Classification: LYASE Ligands: CA, D7K |
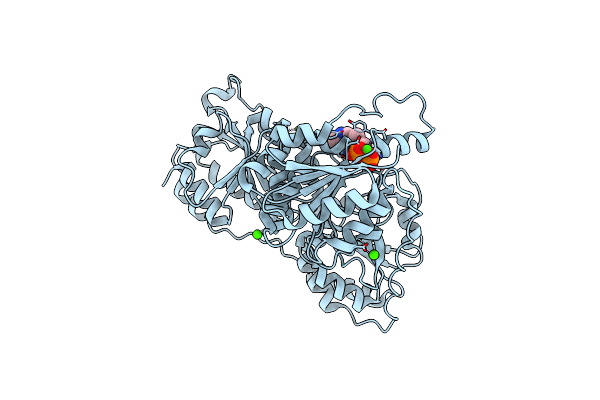 |
Phosphorylation Of An Active Site Serine In A Thdp-Dependent Enzyme By Phosphonate Inactivation
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2006-12-19 Classification: LYASE Ligands: MG, CA, TPP |
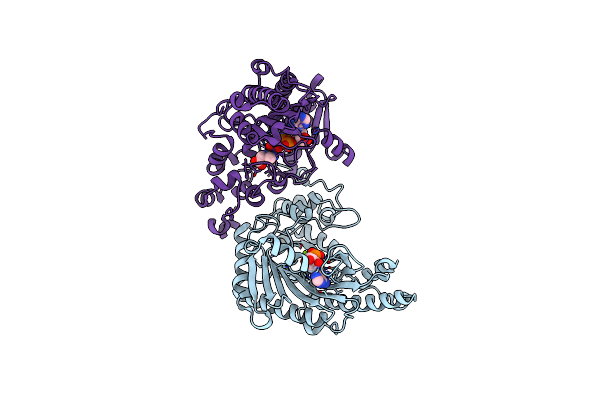 |
The 2.1 Structure Of T. Californica Creatine Kinase Complexed With The Transition-State Analogue Complex, Adp-Mg 2+ /No3-/Creatine
Organism: Torpedo californica
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2005-05-03 Classification: TRANSFERASE Ligands: MG, ADP, NO3, IOM |
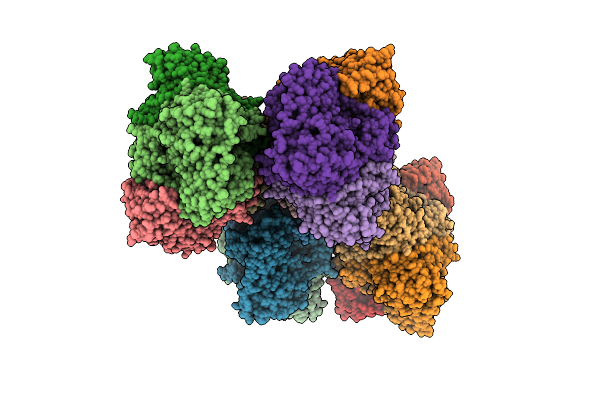 |
Benzoylformate Decarboxylase From Pseudomonas Putida Complexed With An Inhibitor, R-Mandelate
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2003-02-25 Classification: LYASE Ligands: MG, TPP, RMN |
 |
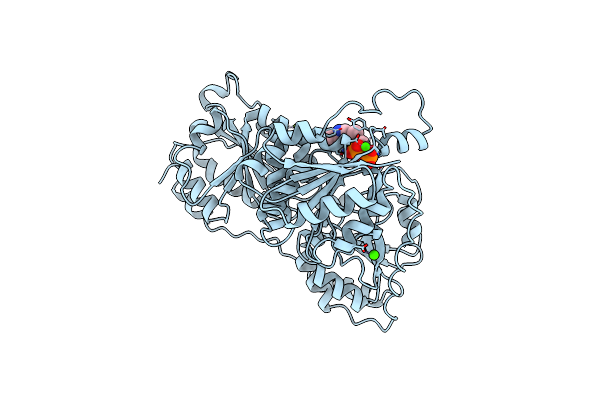
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1998-10-21 Classification: MUCONATE CYCLOISOMERASE |
 |
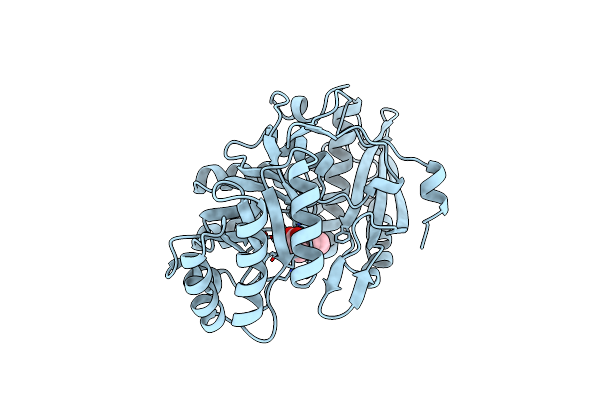
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 1998-06-24 Classification: LYASE Ligands: CA, MG, TPP |
 |
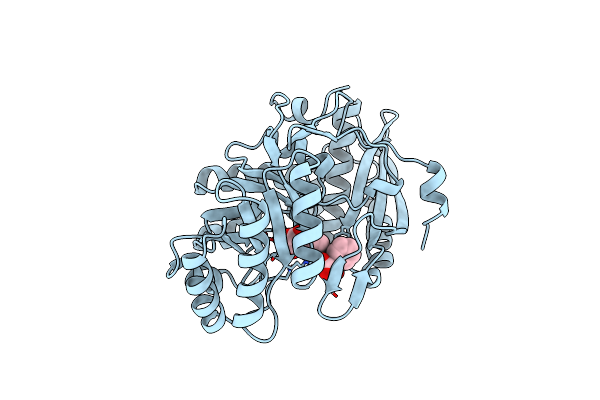
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1996-10-14 Classification: RACEMASE Ligands: MG, APG |
 |
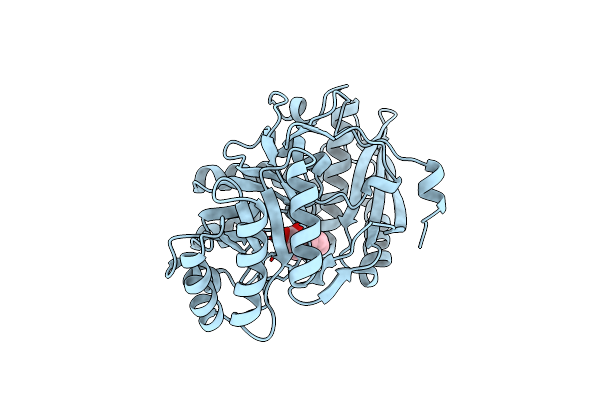
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 1996-10-14 Classification: ISOMERASE Ligands: MG, RMN, SMN |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1996-08-01 Classification: RACEMASE Ligands: MG, APG |
 |
The Role Of Lysine 166 In The Mechanism Of Mandelate Racemase From Pseudomonas Putida: Mechanistic And Crystallographic Evidence For Stereospecific Alkylation By (R)-Alpha-Phenylglycidate
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1994-08-31 Classification: RACEMASE Ligands: MG, APG |
 |
Solution Structure Of A Conserved Dna Sequence From The Hiv-1 Genome: Restrained Molecular Dynamics Simulation With Distance And Torsion Angle Restraints Derived From Two-Dimensional Nmr Spectra
|
 |
Solution Structure Of A Conserved Dna Sequence From The Hiv-1 Genome: Restrained Molecular Dynamics Simulation With Distance And Torsion Angle Restraints Derived From Two-Dimensional Nmr Spectra
|
 |
Solution Structure Of A Conserved Dna Sequence From The Hiv-1 Genome: Restrained Molecular Dynamics Simulation With Distance And Torsion Angle Restraints Derived From Two-Dimensional Nmr Spectra
|
 |
Mechanism Of The Reaction Catalyzed By Mandelate Racemase. 2. Crystal Structure Of Mandelate Racemase At 2.5 Angstroms Resolution: Identification Of The Active Site And Possible Catalytic Residues
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1994-01-31 Classification: RACEMASE Ligands: MN, SO4 |
 |
On The Role Of Lysine 166 In The Mechanism Of Mandelate Racemase From Pseudomonas Putida: Mechanistic And Crystallographic Evidence For Stereospecific Alkylation By (R)-Alpha-Phenylglycidate
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1993-10-31 Classification: RACEMASE Ligands: MG, APG |

