Search Count: 329
 |
Organism: Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: DE NOVO PROTEIN |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of The Bat Rotavirus Apo P[10] Vp8* Receptor Binding Domain At 1.85 Angstrom Resolution
Organism: Rotavirus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-03-26 Classification: VIRAL PROTEIN |
 |
Computationally Designed Tunable C2 Symmetric Tandem Repeat Homodimer, Bound To Cyclic Peptide
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2025-02-05 Classification: DE NOVO PROTEIN |
 |
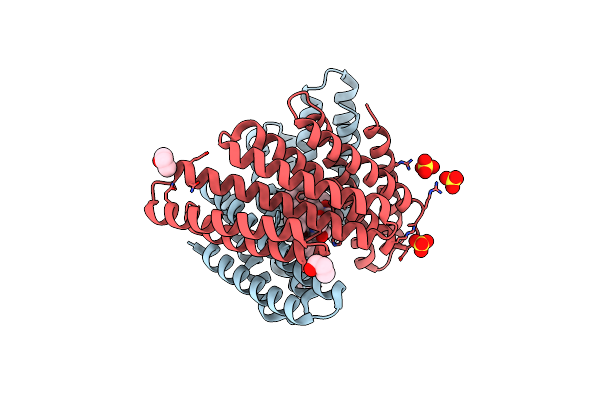
Computationally Designed Tunable C2 Symmetric Tandem Repeat Homodimer, D_3_633_8X Without Peptide
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-12-11 Classification: DE NOVO PROTEIN Ligands: SO4 |
 |
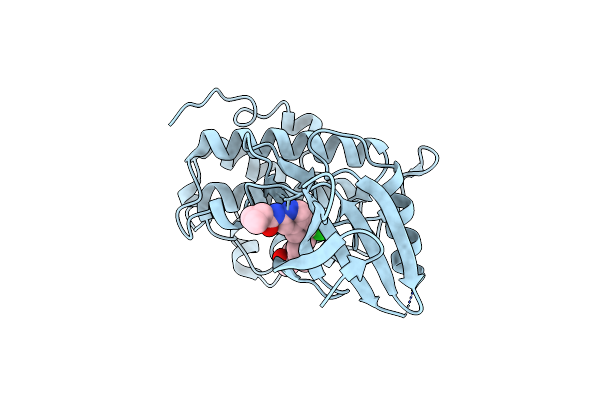
Computationally Designed Tunable C2 Symmetric Tandem Repeat Homodimer, D_3_633_8X Bound To Peptide
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-12-11 Classification: DE NOVO PROTEIN Ligands: POL, SO4 |
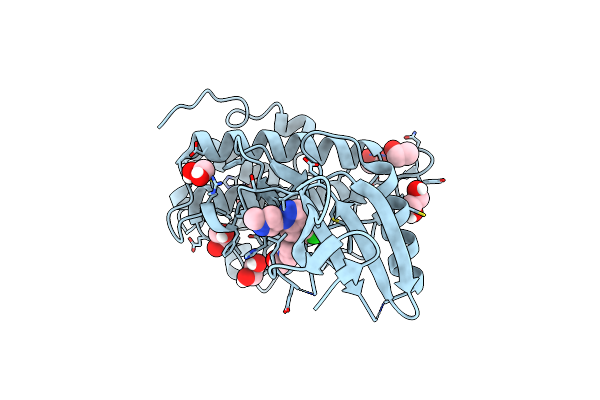 |
Structure Of Chk1 10-Pt. Mutant Complex With Lrrk2 Indazole Inhibitor Compound 6
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2024-09-18 Classification: TRANSFERASE Ligands: A1AV4, EDO |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2024-05-15 Classification: DE NOVO PROTEIN |
 |
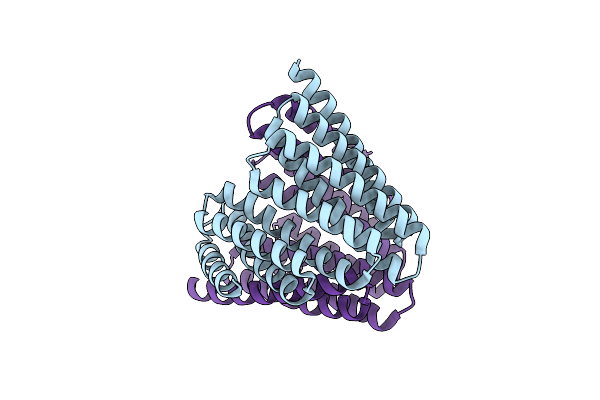
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-03-27 Classification: DE NOVO PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2023-11-01 Classification: TRANSFERASE Ligands: ZXH |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2023-11-01 Classification: TRANSFERASE Ligands: ZXL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-11-01 Classification: TRANSFERASE Ligands: ZXP |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:4.50 Å Release Date: 2023-06-28 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-04-19 Classification: DE NOVO PROTEIN Ligands: EDO |
 |
De Novo Designed Chlorophyll Dimer Protein With Zn Pheophorbide A Methyl Ester, Sp2-Znppam
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-04-19 Classification: DE NOVO PROTEIN Ligands: OE9, PO4, EDO, PGE |
 |
De Novo Designed Chlorophyll Dimer Protein With Zn Pheophorbide A Methyl Ester Matching Geometry Of Purple Bacterial Special Pair, Sp1-Znppam
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-04-19 Classification: DE NOVO PROTEIN Ligands: OE9, SO4, EDO |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2023-04-12 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2023-04-12 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2023-04-12 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2023-04-12 Classification: DE NOVO PROTEIN |

