Search Count: 13
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2023-07-26 Classification: DNA BINDING PROTEIN Ligands: CL, ZQW |
 |
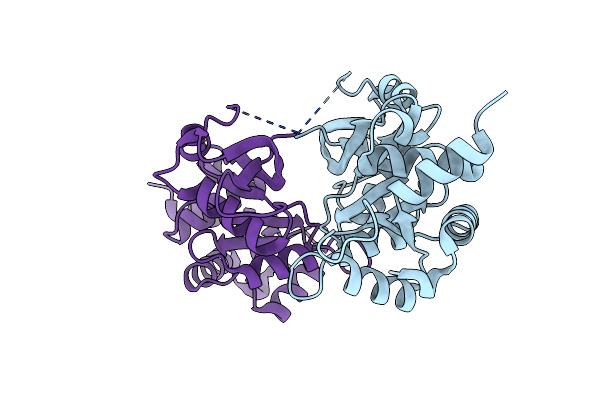
Co-Crystal Structure Of Capcna Bound To The Aoh1996 Derivative, Aoh1996-1Le
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.77 Å Release Date: 2023-07-26 Classification: DNA BINDING PROTEIN Ligands: ZQZ, CL |
 |
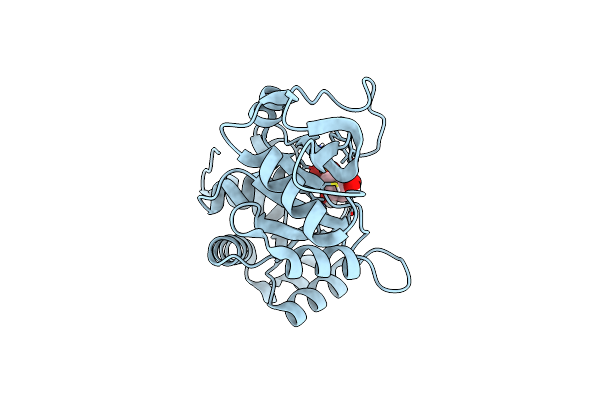
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2022-07-20 Classification: LYASE |
 |
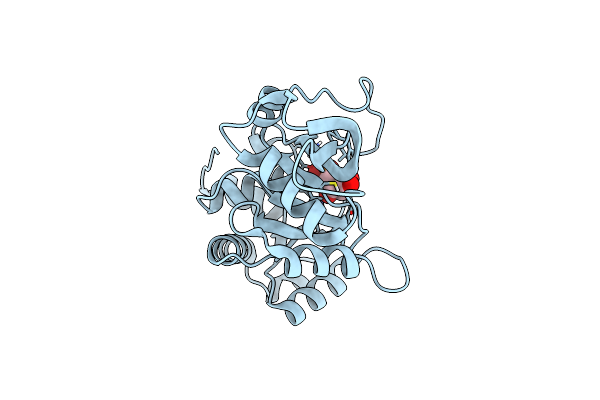
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-07-20 Classification: LYASE Ligands: 5RP |
 |
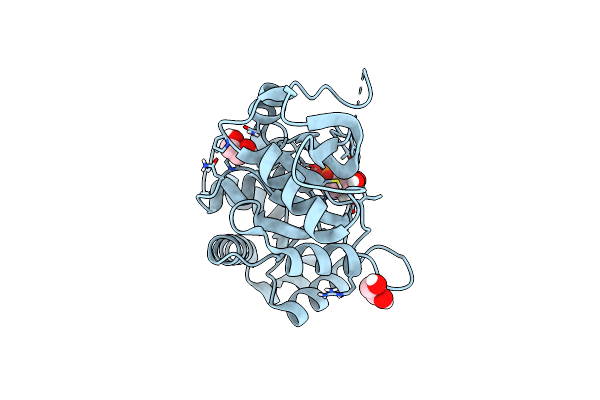
Ribb From Vibrio Cholera Bound With D-Ribose-5-Phosphate (D-R5P) And Manganese
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-07-20 Classification: LYASE Ligands: MN, R5P |
 |
Ribb From Vibrio Cholera Bound With D-Xylulose-5-Phosphate (D-Xy5P) And Manganese
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-07-20 Classification: LYASE Ligands: 5SP, MN, EDO |
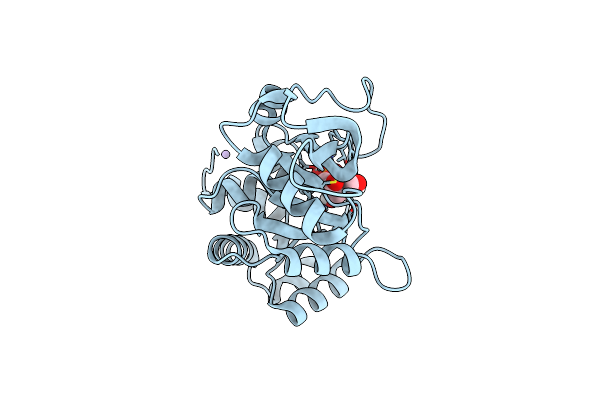 |
Ribb From Vibrio Cholera Bound With L-Xylulose-5-Phosphate (L-Xy5P) And Manganese
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-07-20 Classification: LYASE Ligands: N3U, MN |
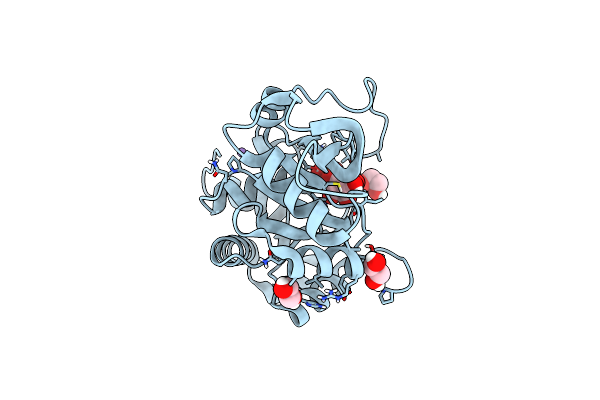 |
Ribb From Vibrio Cholera Bound With Intermediate 1 Of The Reaction Cycle And D-Ribulose-5-Phosphate (D-Ru5P)
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-07-20 Classification: LYASE Ligands: N46, 5RP, MN, EDO |
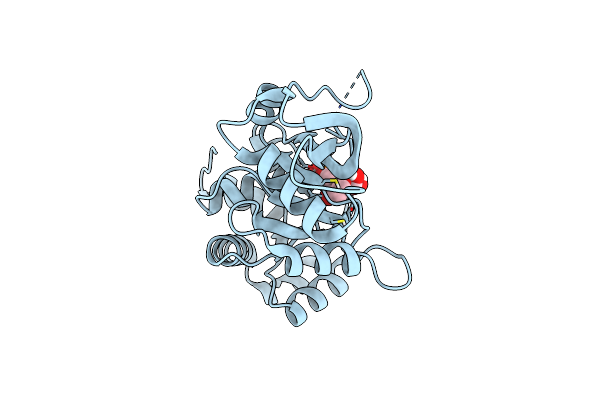 |
Ribb From Vibrio Cholera Bound With Intermediate 2 In The Reaction Cycle And The Products Dhbp And Formate
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-07-20 Classification: LYASE Ligands: N52, N4O, FMT, MN |
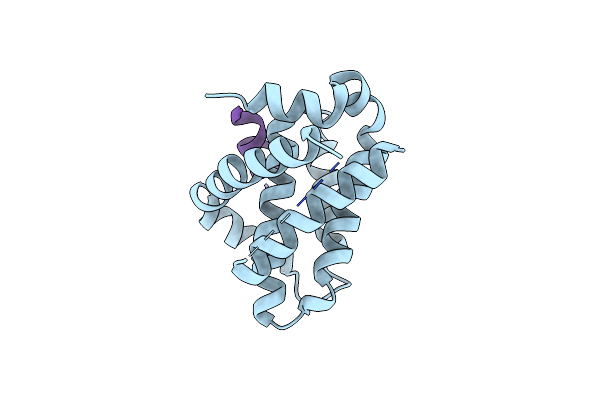 |
Crystal Structure Of Mcl-1 In Complex With 138E12 Peptide, Lys-Covalent Antagonist
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-12-30 Classification: APOPTOSIS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-07-29 Classification: TRANSFERASE Ligands: MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2020-07-29 Classification: TRANSFERASE Ligands: MG |
 |
Pvdf Of Pyoverdin Biosynthesis Is A Structurally Unique N10-Formyltetrahydrofolate-Dependent Formyltransferase
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-02-06 Classification: TRANSFERASE Ligands: FGD, CIT |

