Search Count: 18
 |
Crystal Structure Of Sgvm Methyltransferase In Complex With Alpha-Ketoleucine And Zn2+ Ion
Organism: Streptomyces griseoviridis
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2024-07-24 Classification: TRANSFERASE Ligands: COI, CL, MG, ZN |
 |
Organism: Streptomyces griseoviridis
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2024-07-24 Classification: TRANSFERASE Ligands: ZN, COI, CL, SAM |
 |
Crystal Structure Of Alpha Keto Acid C-Methyl-Transferases Mrsa Native-Form
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2024-07-03 Classification: TRANSFERASE Ligands: MG |
 |
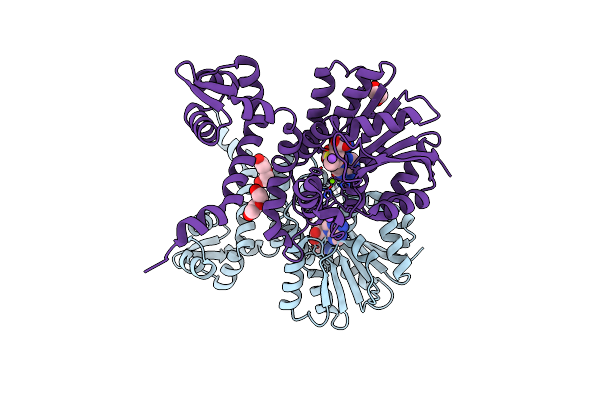
Crystal Structure Of Alpha Keto Acid C-Methyl-Transferases Mrsa Bound To Ketoarginine
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2024-07-03 Classification: TRANSFERASE Ligands: NWG, MG, PEG, PGE, EDO, NA |
 |
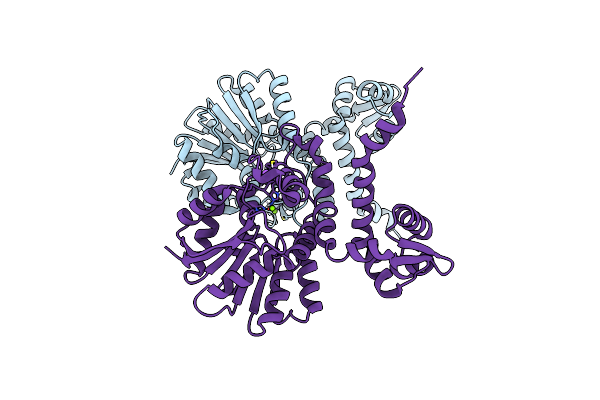
Crystal Structure Of Alpha Keto Acid C-Methyl-Transferases Mrsa Bound To Sam
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2024-07-03 Classification: TRANSFERASE Ligands: MG, SAM, NA, PEG, EDO |
 |
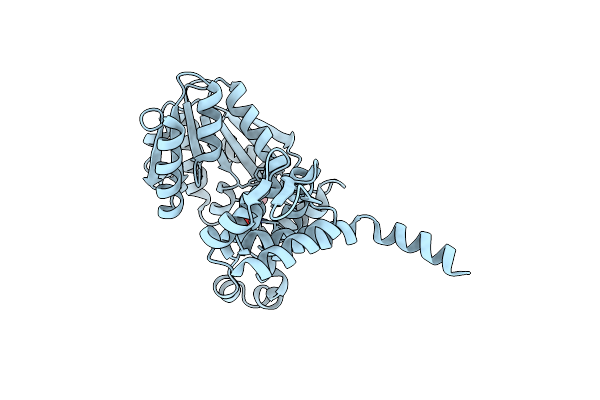
Crystal Structure Of Selenomethionine Derivatized Alpha Keto Acid C-Methyl-Transferases Mrsa
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2024-07-03 Classification: TRANSFERASE Ligands: MG, NA |
 |
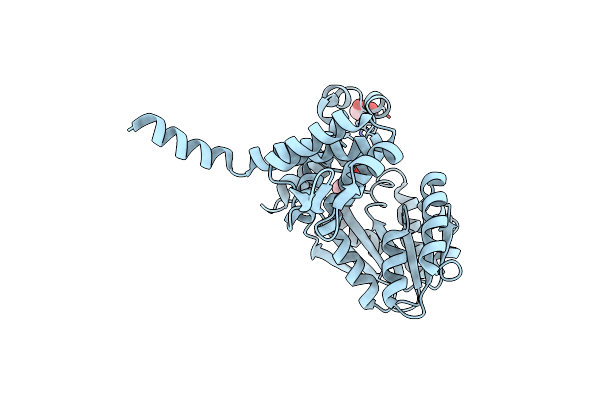
Crystal Structure Of Native Alpha-Keto C-Methyl Transferase Sgvm Bound To Ketoleucine
Organism: Streptomyces griseoviridis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-07-03 Classification: TRANSFERASE Ligands: ZN, COI, CL |
 |
Crystal Structure Of S-Sad Phased Alpha-Keto C-Methyl Transferase Sgvm Bound To Ketoleucine
Organism: Streptomyces griseoviridis
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2024-07-03 Classification: TRANSFERASE Ligands: ZN, COI, CL, GOL |
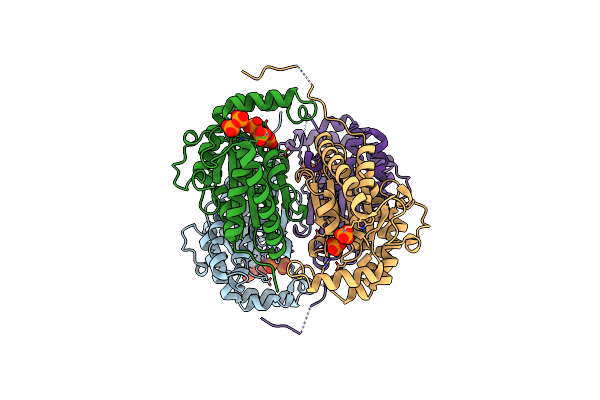 |
Organism: Meiothermus ruber h328
Method: X-RAY DIFFRACTION Release Date: 2018-03-21 Classification: TRANSFERASE Ligands: PO4, DPO |
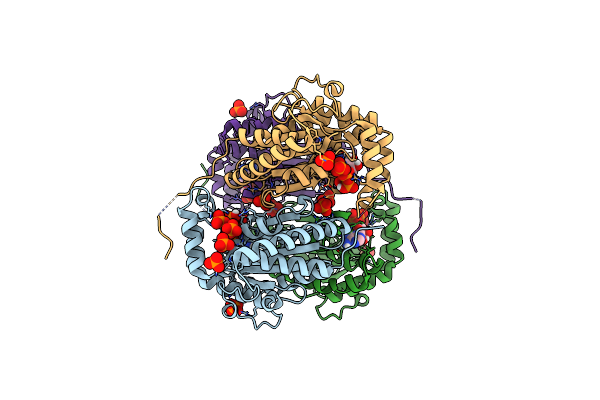 |
Structure Of Polyphosphate Kinase From Meiothermus Ruber N121D Bound To Atp
Organism: Meiothermus ruber h328
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-03-21 Classification: TRANSFERASE Ligands: ATP, PO4 |
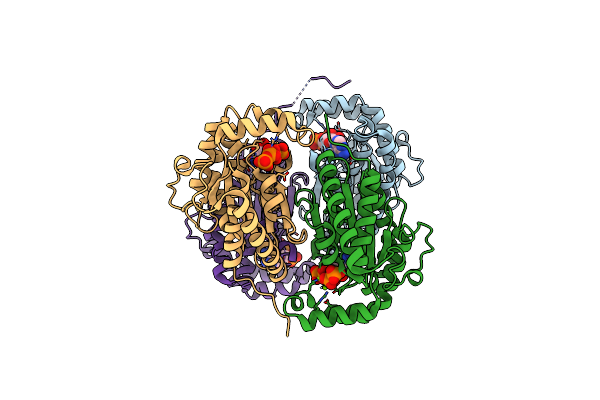 |
Crystal Structure Of Polyphosphate Kinase From Meiothermus Ruber Bound To Adp And Ppi
Organism: Meiothermus ruber h328
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2017-12-20 Classification: TRANSFERASE Ligands: ADP, PPV, MG |
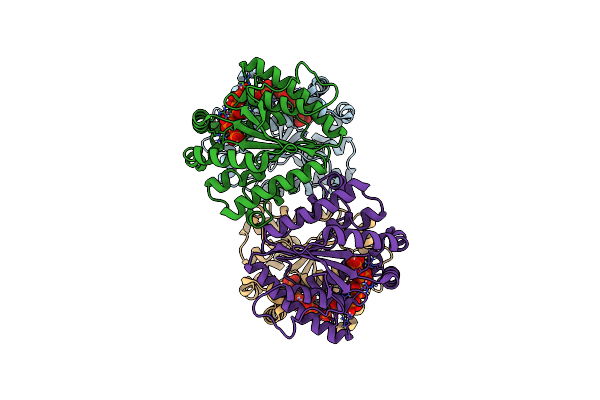 |
Structure Of Polyphosphate Kinase 2 From Francisella Tularensis Schu S4 With Polyphosphate
Organism: Francisella tularensis subsp. tularensis (strain schu s4 / schu 4)
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2017-10-11 Classification: TRANSFERASE Ligands: 9PI |
 |
Structure Of Polyphosphate Kinase 2 From Francisella Tularensis With Amppch2Ppp And Polyphosphate
Organism: Francisella tularensis subsp. tularensis (strain schu s4 / schu 4)
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2017-10-11 Classification: TRANSFERASE Ligands: CL, MG, 6YZ, 6YW |
 |
Structure Of Polyphosphate Kinase 2 Mutant D117N From Francisella Tularensis With Polyphosphate
Organism: Francisella tularensis subsp. tularensis (strain schu s4 / schu 4)
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2017-10-11 Classification: TRANSFERASE Ligands: PO4, 6YX, 6YY, CL, 6YW |
 |
Organism: Meiothermus ruber h328
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-06-21 Classification: TRANSFERASE Ligands: PO4, SO4 |
 |
Organism: Meiothermus ruber h328
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2017-06-21 Classification: TRANSFERASE Ligands: AMP, PO4, SO4, MG |
 |
Crystal Structure Of Polyphosphate Kinase From Meiothermus Ruber Bound To Atp
Organism: Meiothermus ruber
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2017-06-21 Classification: TRANSFERASE Ligands: ATP, MG, SO4, GOL |
 |
Crystal Structure Of Polyphosphate Kinase From Meiothermus Ruber Bound To Adp
Organism: Meiothermus ruber h328
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-06-21 Classification: TRANSFERASE Ligands: ADP, PO4, MG, GOL, CL |

