Search Count: 26
 |
Bifunctional Non-Haem Iron Oxygenase Novr Involved In Novobiocin Biosynthesis With Product Bound
Organism: Streptomyces niveus
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: LYASE Ligands: A1JKI, FE, SO4, EDO |
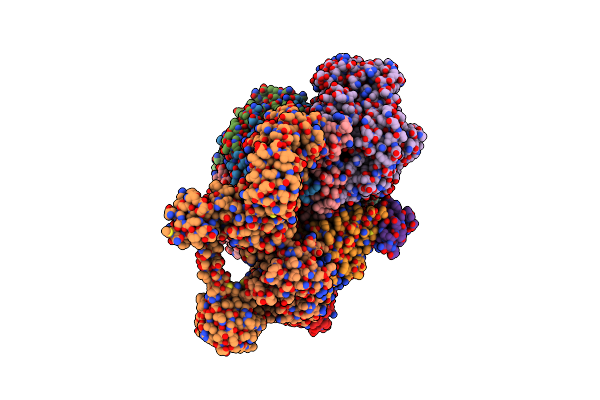 |
Organism: Methanococcus maripaludis
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: SHT, TP7, COM, F43, S5Q, ZN, ATP, MG |
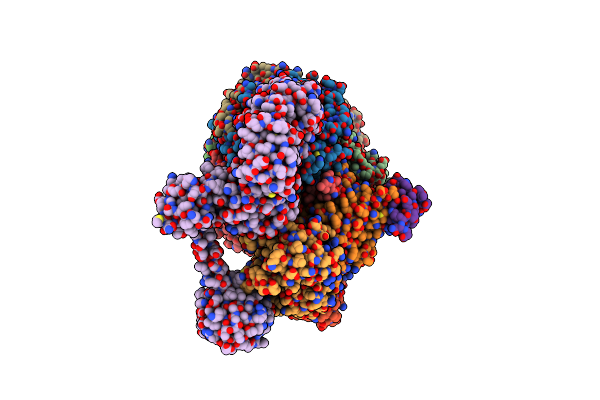 |
Organism: Methanococcus maripaludis
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: F43, SHT, COM, TP7, S5Q |
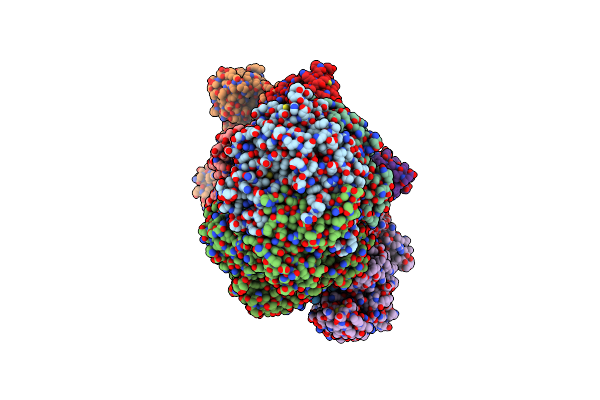 |
Methyl-Coenzyme M Reductase Activation Complex Binding To The A2 Component After Incubation With Atp
Organism: Methanococcus maripaludis
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: F43, COM, TP7, SHT, S5Q, ZN, ATP, MG |
 |
Cryo-Em Structure Of A Nautilus-Like Hflk/C Assembly In Complex With Ftsh Aaa Protease
Organism: Escherichia coli bl21
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: CHAPERONE |
 |
Organism: Sulfurospirillum multivorans
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2019-06-05 Classification: LYASE |
 |
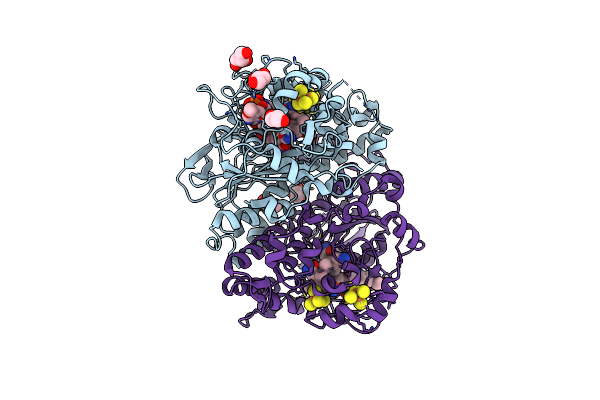
Photo-Driven Hydrogen Evolution By An Artificial Hydrogenase Utilizing The Biotin-Streptavidin Technology
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-03-13 Classification: OXIDOREDUCTASE Ligands: 9CO |
 |
Pce Reductive Dehalogenase From S. Multivorans With 5-Methoxybenzimidazolyl-Norcobamide Cofactor
Organism: Sulfurospirillum multivorans dsm 12446
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-02-14 Classification: OXIDOREDUCTASE Ligands: SF4, OBL, BEN, GOL |
 |
Pce Reductive Dehalogenase From S. Multivorans With 6-Hydroxybenzimidazolyl Norcobamide Cofactor
Organism: Sulfurospirillum multivorans
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2018-02-14 Classification: OXIDOREDUCTASE Ligands: SF4, 9QQ, BEN, GOL |
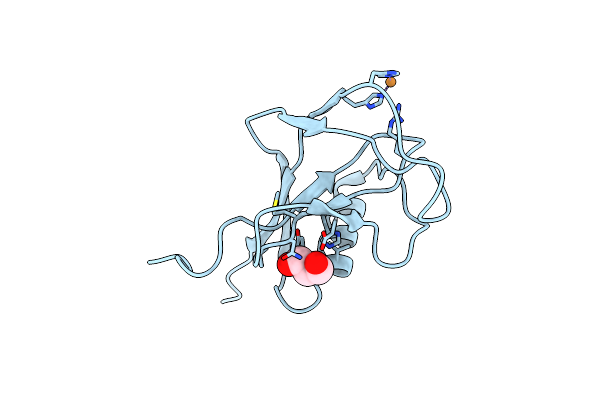 |
Crystal Structure Of The N-Terminal, Growth Factor-Like Domain Of The Amyloid Precursor Protein Bound To Copper
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2014-07-23 Classification: METAL BINDING PROTEIN Ligands: CU, GOL |
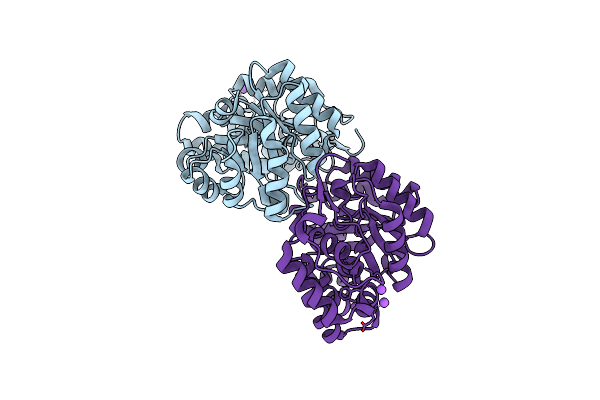 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-07-18 Classification: LYASE |
 |
The Structure Of Dihydrodipicolinate Synthase 2 From Arabidopsis Thaliana In Complex With (S)-Lysine
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-07-18 Classification: LYASE |
 |
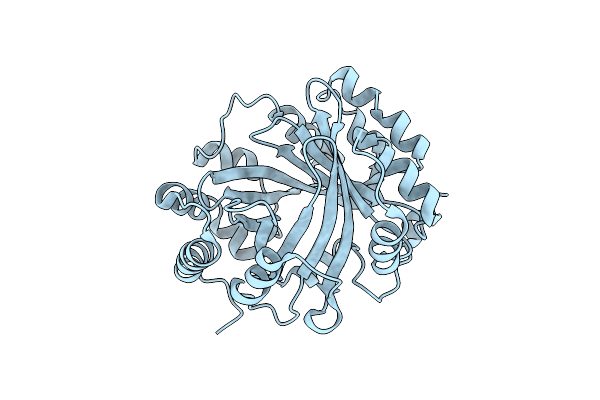
Structural And Mechanistic Analysis Of The Magnesium-Independent Aromatic Prenyltransferase Cloq From The Clorobiocin Biosynthetic Pathway
Organism: Streptomyces roseochromogenes subsp. oscitans
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2010-10-27 Classification: TRANSFERASE Ligands: FMT, 34H |
 |
Structural And Mechanistic Analysis Of The Magnesium-Independent Aromatic Prenyltransferase Cloq From The Clorobiocin Biosynthetic Pathway
Organism: Streptomyces roseochromogenes subsp. oscitans
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2010-10-27 Classification: TRANSFERASE |
 |
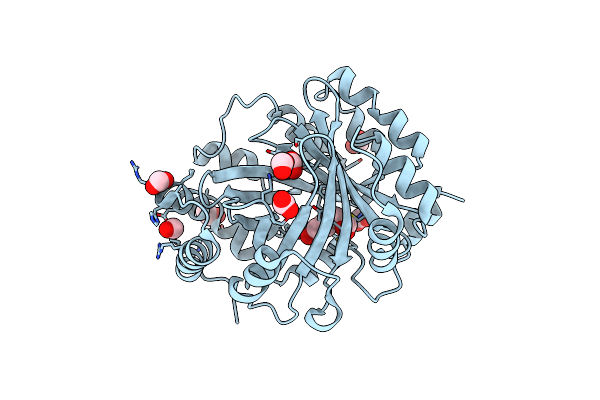
Structural And Mechanistic Analysis Of The Magnesium-Independent Aromatic Prenyltransferase Cloq From The Clorobiocin Biosynthetic Pathway
Organism: Streptomyces roseochromogenes subsp. oscitans
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2010-10-27 Classification: TRANSFERASE Ligands: EDO, FMT |
 |
Structural And Mechanistic Analysis Of The Magnesium-Independent Aromatic Prenyltransferase Cloq From The Clorobiocin Biosynthetic Pathway
Organism: Streptomyces roseochromogenes subsp. oscitans
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2010-10-27 Classification: TRANSFERASE Ligands: 34H, EDO, FMT |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2010-04-07 Classification: PLANT PROTEIN Ligands: SO4, EDO |
 |
Crystal Structures Of The Gach Receptor Of Streptomyces Glaucescens Gla.O In The Unliganded Form And In Complex With Acarbose And An Acarbose Homolog. Comparison With Acarbose-Loaded Maltose Binding Protein Of Salmonella Typhimurium.
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2010-02-16 Classification: TRANSPORT PROTEIN |
 |
Crystal Structures Of The Gach Receptor Of Streptomyces Glaucescens Gla.O In The Unliganded Form And In Complex With Acarbose And An Acarbose Homolog. Comparison With Acarbose-Loaded Maltose Binding Protein Of Salmonella Typhimurium.
Organism: Streptomyces glaucescens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2010-02-16 Classification: TRANSPORT PROTEIN Ligands: SO4 |
 |
Crystal Structures Of The Gach Receptor Of Streptomyces Glaucescens Gla.O In The Unliganded Form And In Complex With Acarbose And An Acarbose Homolog. Comparison With Acarbose-Loaded Maltose Binding Protein Of Salmonella Typhimurium.
Organism: Streptomyces glaucescens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2010-02-16 Classification: TRANSPORT PROTEIN |

