Search Count: 18
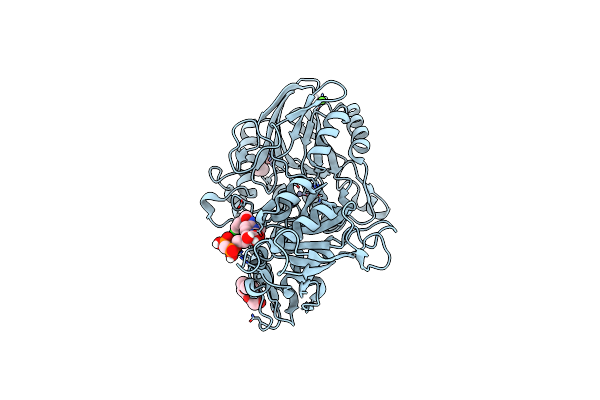 |
Human Cd73 (Ecto 5'-Nucleotidase) In Complex With Mrs4598 (A 3-Methyl-Cmpcp Derivative, Compound 16 In Paper) In The Open State
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-02-16 Classification: HYDROLASE Ligands: ZN, CA, BOI, PG4 |
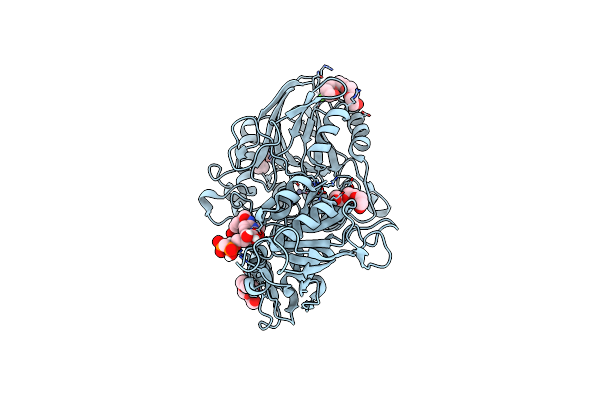 |
Human Cd73 (Ecto 5'-Nucleotidase) In Complex With Mrs4602 (A 3-Methyl-Cmpcp Derivative, Compound 21 In Paper) In The Open State
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-02-16 Classification: HYDROLASE Ligands: ZN, CA, BW0, 1PE |
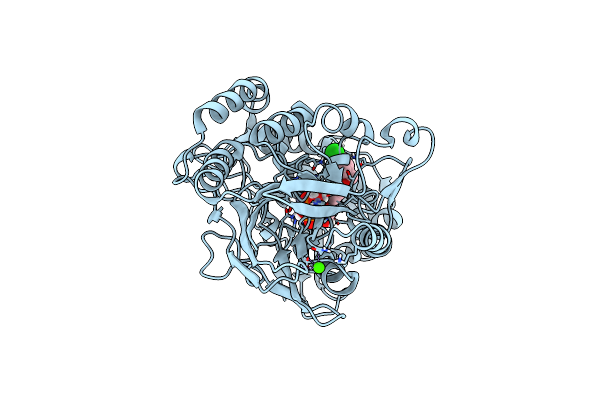 |
Human Cd73 (Ecto 5'-Nucleotidase) In Complex With Mrs4598 (A 3-Methyl-Cmpcp Derivative, Compound 16 In Paper) In The Closed State (Crystal Form Iii)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2022-02-16 Classification: HYDROLASE Ligands: ZN, CA, BOI |
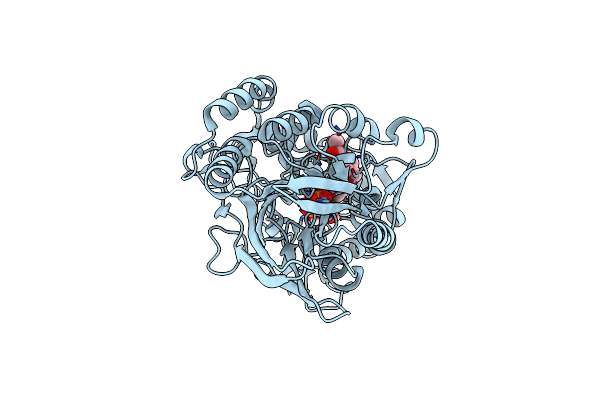 |
Human Cd73 (Ecto 5'-Nucleotidase) In Complex With Mrs4602 (A 3-Methyl-Cmpcp Derivative, Compound 21 In Paper) In The Closed State (Crystal Form Iii)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2022-02-16 Classification: HYDROLASE Ligands: ZN, BW0 |
 |
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-10-13 Classification: TRANSFERASE Ligands: SO4, NHE |
 |
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2010-10-13 Classification: TRANSFERASE |
 |
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2010-10-13 Classification: TRANSFERASE Ligands: GOL, NHE, PO4 |
 |
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2010-10-13 Classification: TRANSFERASE Ligands: NHE, GOL, SO4 |
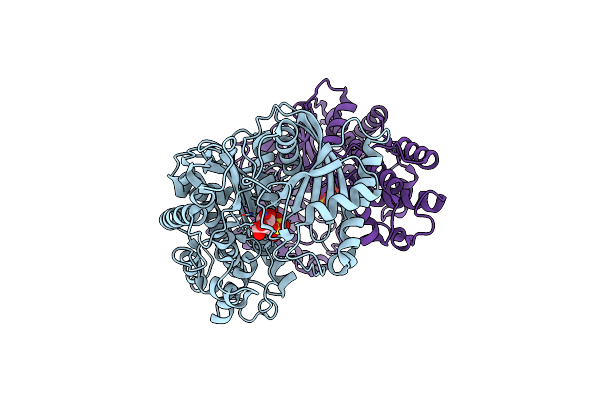 |
Crystal Structure Of Dimeric Klhxk1 In Crystal Form Vii With Glucose Bound (Open State)
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2010-10-13 Classification: TRANSFERASE Ligands: BGC, SO4 |
 |
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2010-10-13 Classification: TRANSFERASE Ligands: GOL, PO4 |
 |
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2010-10-13 Classification: TRANSFERASE Ligands: ANP |
 |
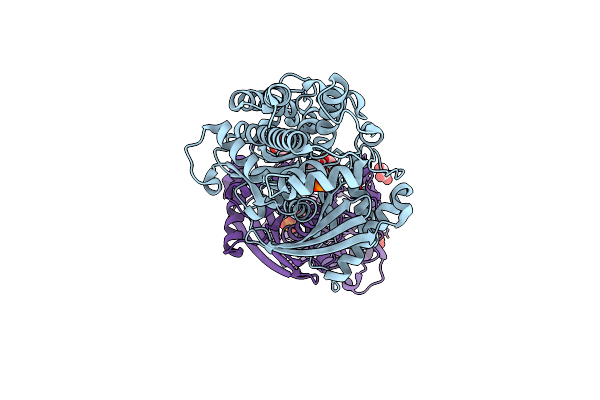
Crystal Structure Of Monomeric Klhxk1 In Crystal Form Xi With Glucose Bound (Closed State)
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2010-10-13 Classification: TRANSFERASE Ligands: GLC, BGC, CL |
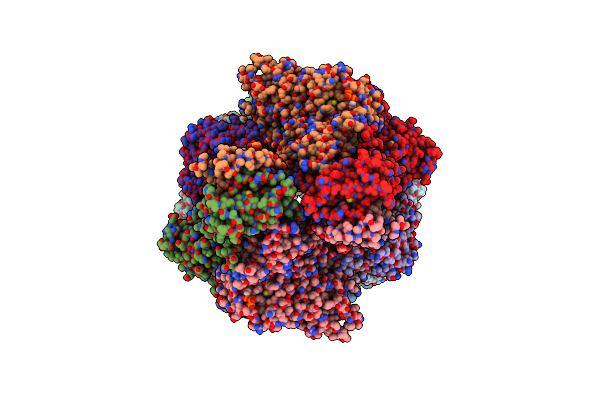 |
Organism: Pichia pastoris
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2010-10-06 Classification: TRANSFERASE Ligands: SO4, ATP |
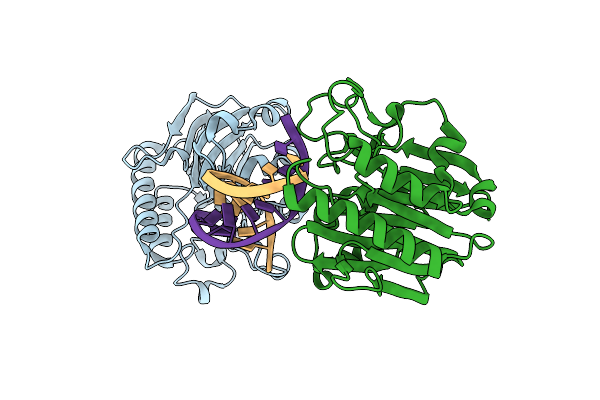 |
Organism: Archaeoglobus fulgidus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2008-12-09 Classification: LYASE |
 |
Organism: Bordetella bronchiseptica
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2008-03-18 Classification: LYASE Ligands: BME, PO4, EDO |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2007-10-30 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2007-10-30 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2007-10-30 Classification: OXIDOREDUCTASE |

