Search Count: 26
 |
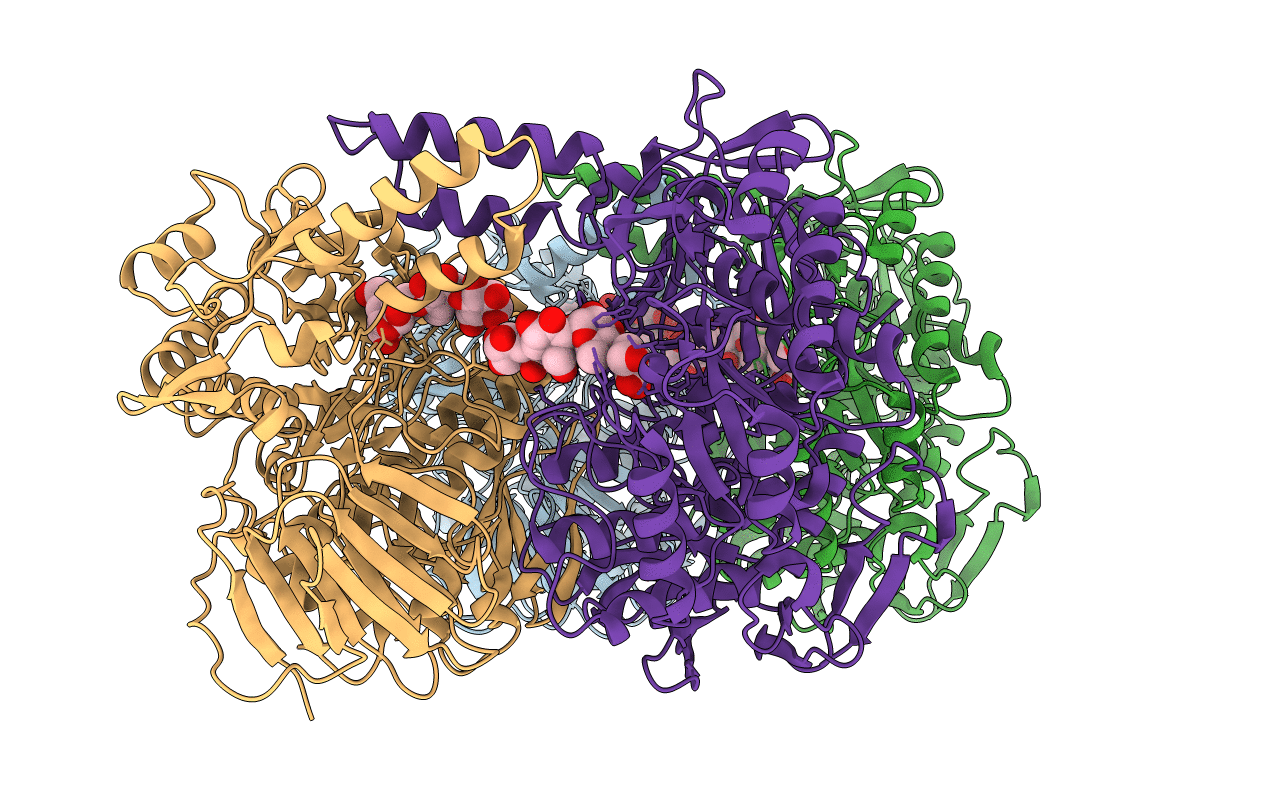
The Crystal Structure Of The D307A Mutant Of Alpha-Glucosidase (Family 31) From Ruminococcus Obeum Atcc 29174 In Complex With Acarbose
Organism: Ruminococcus obeum
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2011-01-26 Classification: hydrolase/hydrolase inhibitor Ligands: GLC, AC1, GOL |
 |
The Crystal Structure Of The W169Y Mutant Of Alpha-Glucosidase (Gh31 Family) From Ruminococcus Obeum Atcc 29174 In Complex With Acarbose
Organism: Blautia obeum atcc 29174
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2010-11-24 Classification: hydrolase/hydrolase inhibitor |
 |
The Crystal Structure Of A Fat Acid (Stearic Acid)-Binding Protein From Eubacterium Ventriosum Atcc 27560.
Organism: Eubacterium ventriosum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-09-22 Classification: LIPID BINDING PROTEIN Ligands: STE |
 |
The Crystal Structure Of The W169Y Mutant Of Alpha-Glucosidase (Family 31) From Ruminococcus Obeum Atcc 29174
Organism: Ruminococcus obeum
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2010-07-28 Classification: HYDROLASE Ligands: GOL |
 |
The Crystal Structure Of The The Crystal Structure Of The D420A Mutant Of The Alpha-Glucosidase (Family 31) From Ruminococcus Obeum Atcc 29174
Organism: Ruminococcus obeum
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2010-07-21 Classification: HYDROLASE Ligands: TRS |
 |
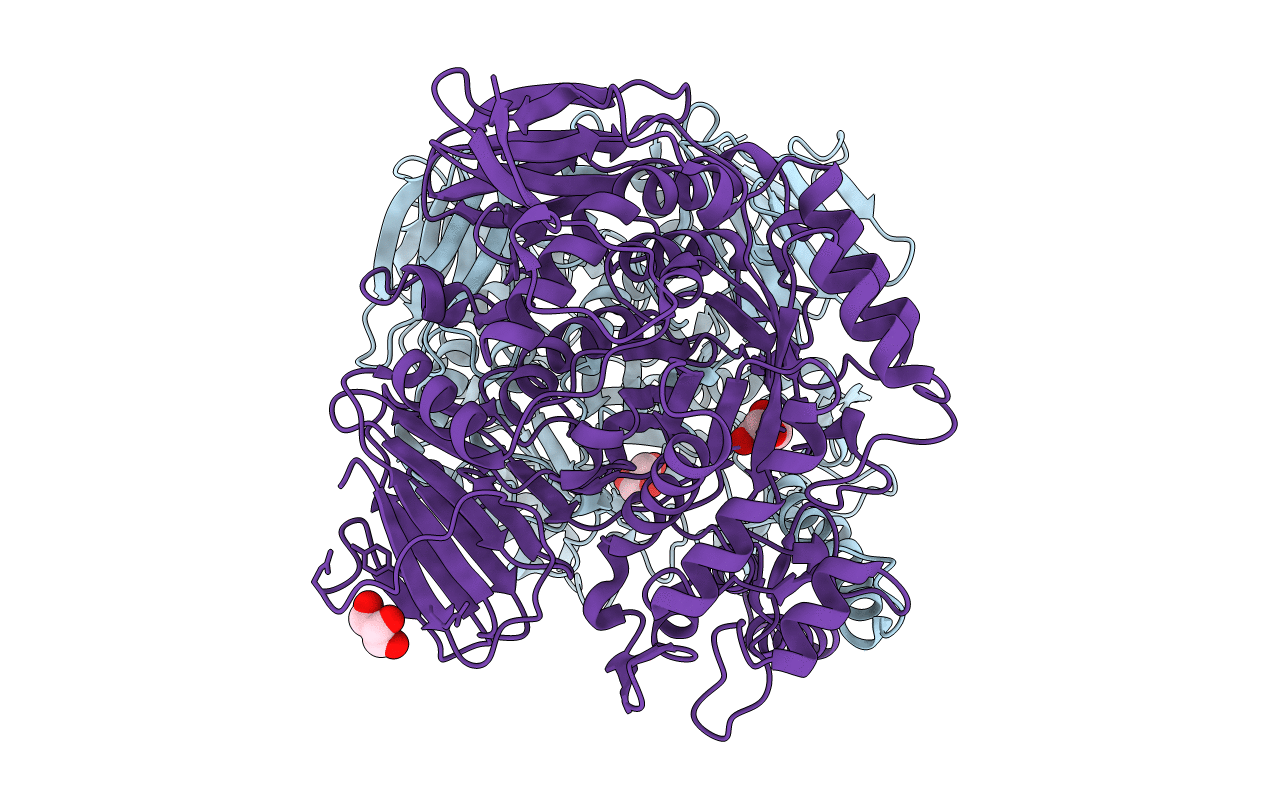
The Crystal Structure Of The D307A Mutant Of Glycoside Hydrolase (Family 31) From Ruminococcus Obeum Atcc 29174 In Complex With Isomaltose
Organism: Ruminococcus obeum
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2010-06-23 Classification: HYDROLASE Ligands: GLC, BGC |
 |
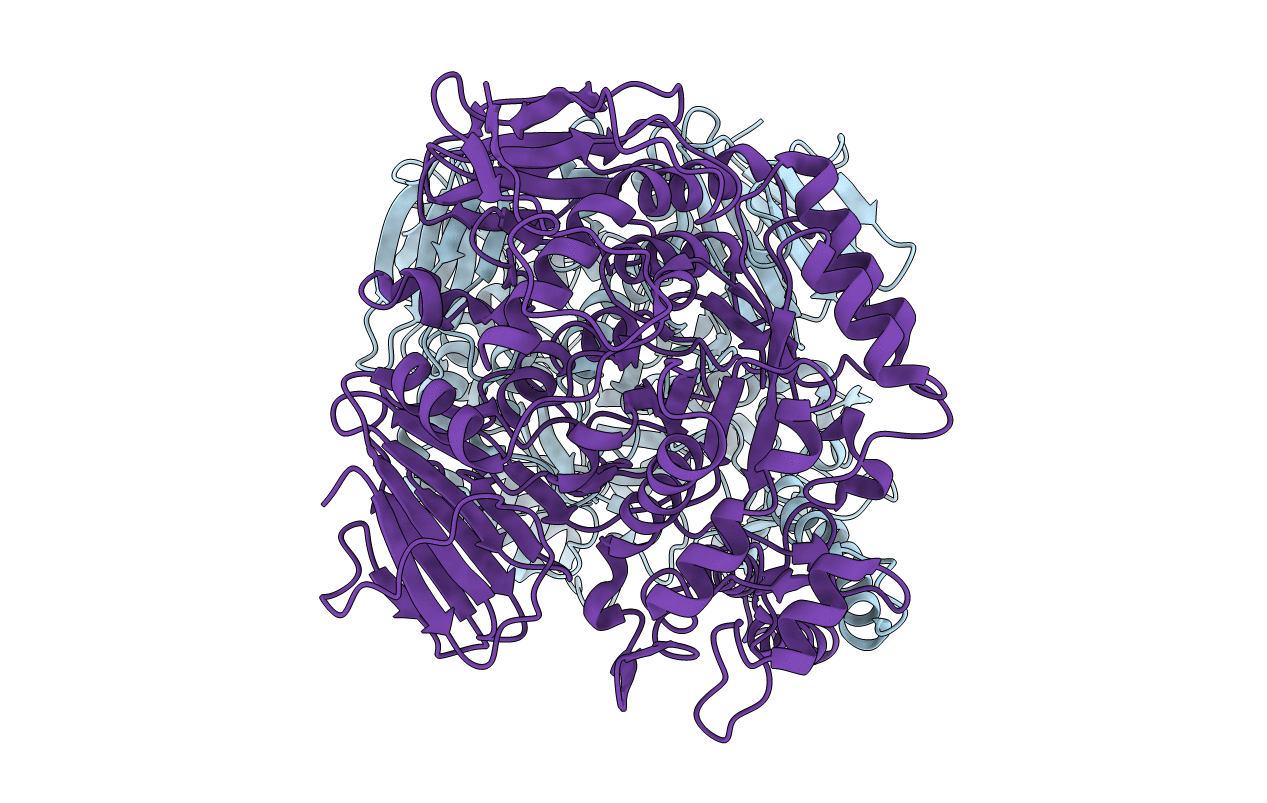
The Crystal Structure Of The Alpha-Glucosidase (Family 31) From Ruminococcus Obeum Atcc 29174
Organism: Ruminococcus obeum
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2010-06-23 Classification: HYDROLASE Ligands: GOL |
 |
The Crystal Structure Of The D307A Mutant Of Glycoside Hydrolase (Family 31) From Ruminococcus Obeum Atcc 29174
Organism: Ruminococcus obeum
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2010-04-21 Classification: HYDROLASE |
 |
The Crystal Structure Of The D73A Mutant Of Glycoside Hydrolase (Family 31) From Ruminococcus Obeum Atcc 29174
Organism: Ruminococcus obeum
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2010-03-23 Classification: HYDROLASE Ligands: GOL |
 |
Crystal Structure Of Probable Methyltransferase From Bordetella Pertussis Tohama I
Organism: Bordetella pertussis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2009-12-08 Classification: TRANSFERASE Ligands: GOL, SO4 |
 |
The Crystal Structure Of One Domain Of The Conserved Protein From Methanosarcina Mazei Go1
Organism: Methanosarcina mazei
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-10-20 Classification: structural genomics, unknown function |
 |
Crystal Structure Of Putative Zn-Dependent Alcohol Dehydrogenases From Rhodobacter Sphaeroides.
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2009-09-15 Classification: OXIDOREDUCTASE Ligands: SAM, CL |
 |
The Crystal Structure Of The Inactive Peptidase Domain Of A Putative Zinc Protease From Bordetella Parapertussis To 2.2A
Organism: Bordetella parapertussis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-09-08 Classification: HYDROLASE Ligands: PEG |
 |
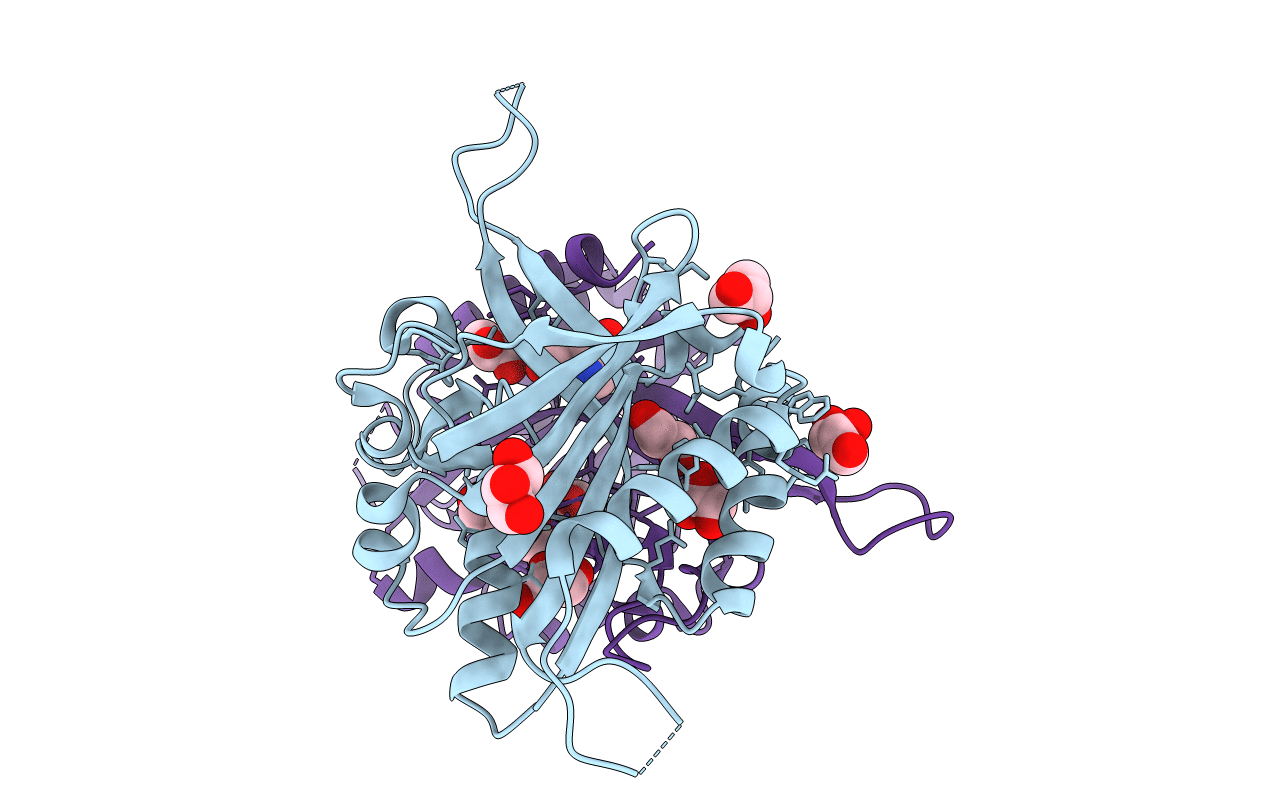
The Crystal Structure Of A Putative Phenylalanyl-Trna Synthetase (Phers) Beta Chain Domain From Bacteroides Fragilis To 2.1A
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2009-09-01 Classification: LIGASE Ligands: MG |
 |
The Crystal Structure Of The Beta Subunit Of A Phenylalanyl-Trna Synthetase From Porphyromonas Gingivalis W83
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2009-07-28 Classification: LIGASE Ligands: GOL, TAM |
 |
Crystal Structure Of A Basic Coiled-Coil Protein Of Unknown Function From Eubacterium Eligens Atcc 27750
Organism: Eubacterium eligens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-07-14 Classification: structural genomics, unknown function Ligands: IOD, GOL |
 |
Crystal Structure Of Uncharacterized Protein Conserved In Bacteria Duf2179 From Eubacterium Ventriosum
Organism: Eubacterium ventriosum
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2009-07-07 Classification: structural genomics, unknown function |
 |
Crystal Structure Of Putative Precorrin-6Y C5,15-Methyltransferase Targeted Domain From Corynebacterium Diphtheriae
Organism: Corynebacterium diphtheriae
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2009-07-07 Classification: TRANSFERASE Ligands: ACY, MG, CA |
 |
The Crystal Structure Of A Thioredoxin-Like Oxidoreductase From Silicibacter Pomeroyi Dss-3
Organism: Silicibacter pomeroyi
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2009-06-09 Classification: OXIDOREDUCTASE Ligands: SO4, EDO |
 |
Crystal Structure Of A Probable Rna-Binding Protein From Clostridium Symbiosum Atcc 14940
Organism: Clostridium symbiosum atcc 14940
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2009-03-31 Classification: RNA BINDING PROTEIN |

