Search Count: 26
 |
Crystal Structure Of Sars-Cov-2 Receptor Binding Domain In Complex With Antibodies Ecr3022.20 And Cc12.3
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2023-08-23 Classification: IMMUNE SYSTEM/VIRAL PROTEIN Ligands: NAG |
 |
Crystal Structure Of Sars-Cov-2 Spike Protein Receptor-Binding Domain In Complex With Antibody Cc12.1 Fab And Nanobody Nb-C4-225
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2023-07-05 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Crystal Structure Of Sars-Cov-2 Spike Protein Receptor-Binding Domain In Complex With Antibody Cc12.1 Fab And Nanobody Nb-C4-240
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2023-07-05 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Crystal Structure Of Sars-Cov-2 Spike Protein Receptor-Binding Domain In Complex With Antibody Cc12.1 Fab And Nanobody Nb-C4-255
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2023-07-05 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Lm18/Nb136 Bispecific Tetra-Nanobody Immunoglobulin In Complex With Sars-Cov-2-6P-Mut7 S Protein (Focused Refinement)
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2022-08-24 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2022-08-24 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Cc6.33 Igg In Complex With Sars-Cov-2-6P-Mut7 S Protein (Non-Uniform Refinement)
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2022-08-24 Classification: VIRAL PROTEIN/Immune System Ligands: NAG |
 |
Cc6.33 Igg In Complex With Sars-Cov-2-6P-Mut7 S Protein (Rbd/Fv Local Refinement)
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2022-08-24 Classification: VIRAL PROTEIN/Immune System Ligands: NAG |
 |
Cc6.30 Fragment Antigen Binding In Complex With Sars-Cov-2-6P-Mut7 S Protein (Non-Uniform Refinement)
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-08-24 Classification: VIRAL PROTEIN/Immune System Ligands: NAG |
 |
Cc6.30 Fragment Antigen Binding In Complex With Sars-Cov-2-6P-Mut7 S Protein (Rbd/Fv Local Refinement)
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2022-08-24 Classification: VIRAL PROTEIN/Immune System Ligands: NAG |
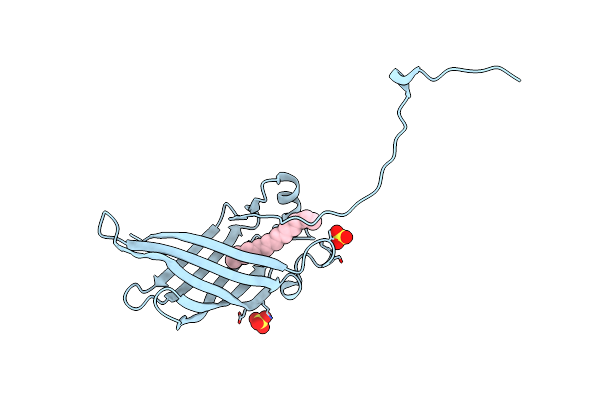 |
Crystal Structure Of Campylobacter Jejuni Ycei Protein That Crystallizes With Large Solvent Channels For Nanotechnology Applications
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2018-01-03 Classification: UNKNOWN FUNCTION Ligands: LFA, SO4 |
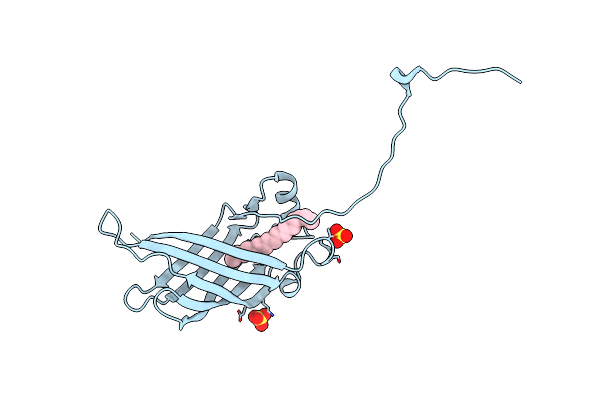 |
Crystal Structure Of Mutant Cj Ycei Protein (Cj-G34C) For Nanotechnology Applications
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-01-03 Classification: UNKNOWN FUNCTION Ligands: UNL, SO4 |
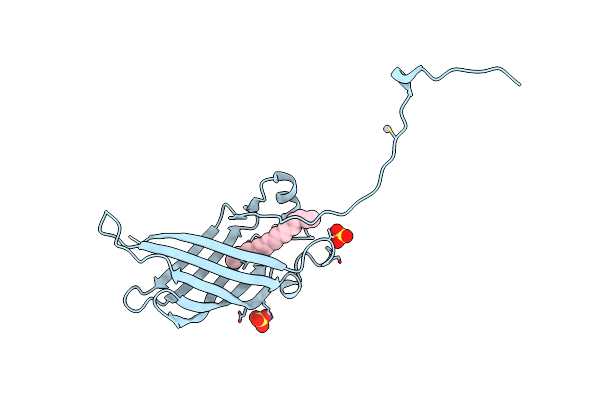 |
Crystal Structure Of Mutant Cj Ycei Protein (Cj-G34C) With Hydroxymercuribenzoic Acid Guest Structure
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2018-01-03 Classification: UNKNOWN FUNCTION Ligands: UNL, SO4, MBO |
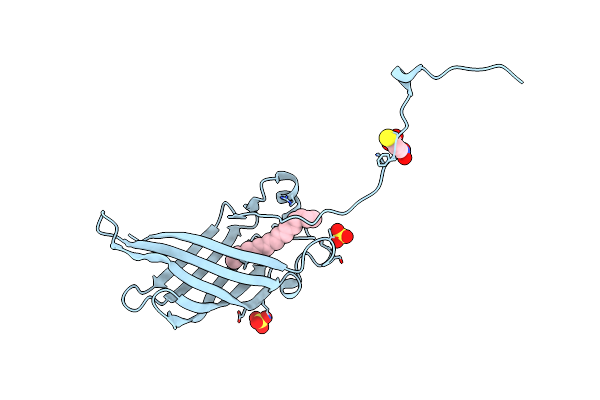 |
Crystal Structure Of Mutant Cj Ycei Protein (Cj-G34C) With 5-Mercapto-2-Nitrobenzoic Acid Guest Structure
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2018-01-03 Classification: UNKNOWN FUNCTION Ligands: UNL, SO4, MNB |
 |
Crystal Structure Of Mutant Cj Ycei Protein (Cj-G34C) With Selenocysteine Guest Structure
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2018-01-03 Classification: UNKNOWN FUNCTION Ligands: UNL, SO4, SEC |
 |
Crystal Structure Of Mutant Cj Ycei Protein (Cj-N48C) For Nanotechnology Applications
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2018-01-03 Classification: UNKNOWN FUNCTION Ligands: UNL, SO4 |
 |
Crystal Structure Of Mutant Cj Ycei Protein (Cj-N48C) With 5-Mercapto-2-Nitrobenzoic Acid Guest Structure
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-01-03 Classification: UNKNOWN FUNCTION Ligands: UNL, SO4, MNB |
 |
Crystal Structure Of Mutant Cj Ycei Protein (Cj-N48C) With Monobromobimane Guest Structure
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2018-01-03 Classification: UNKNOWN FUNCTION Ligands: UNL, SO4, 9UM |
 |
Crystal Structure Of Mutant Cj Ycei Protein (Cj-N48C) With Mercuribenzoic Acid Guest Structure
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2018-01-03 Classification: UNKNOWN FUNCTION Ligands: UNL, SO4, MBO |

