Search Count: 573
 |
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: ANTIMICROBIAL PROTEIN |
 |
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: ANTIMICROBIAL PROTEIN |
 |
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: ANTIMICROBIAL PROTEIN Ligands: MER |
 |
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: ANTIMICROBIAL PROTEIN Ligands: NXL |
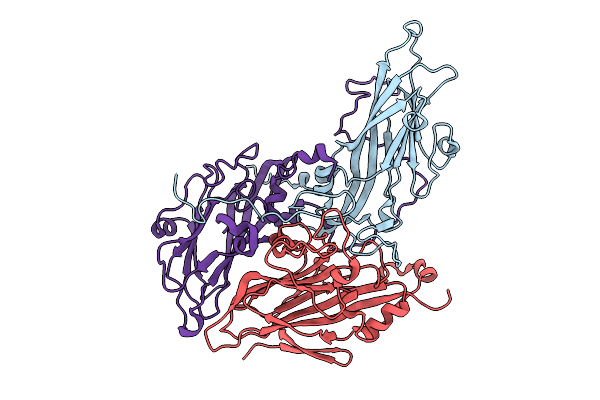 |
Organism: Enterovirus b75
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN |
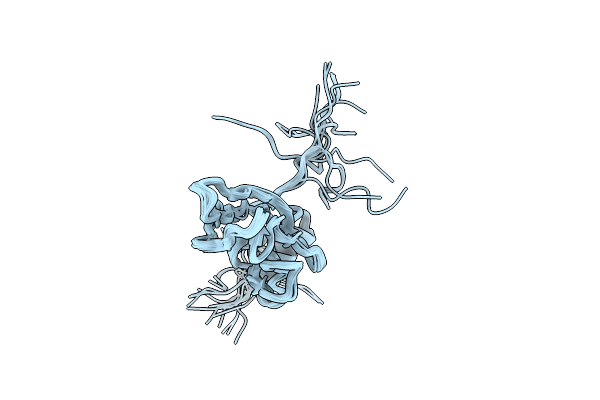 |
A Conserved Beta-Sandwich Fold Is Required For Secretion Of Lipoproteins By A Novel Type I Secretion System
Organism: Escherichia coli etec h10407
Method: SOLUTION NMR Release Date: 2025-12-03 Classification: TRANSPORT PROTEIN |
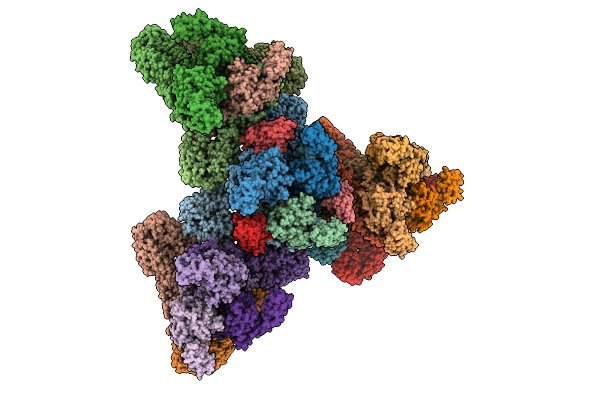 |
Venezuelan Equine Encephalitis Virus In Complex With The Single Domain Antibody V2B3
Organism: Venezuelan equine encephalitis virus, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: Viral protein/Immune System |
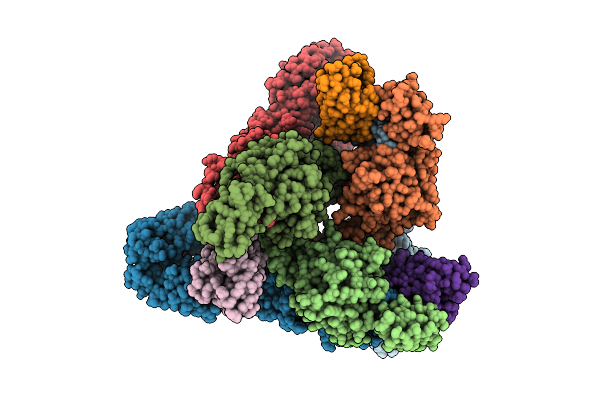 |
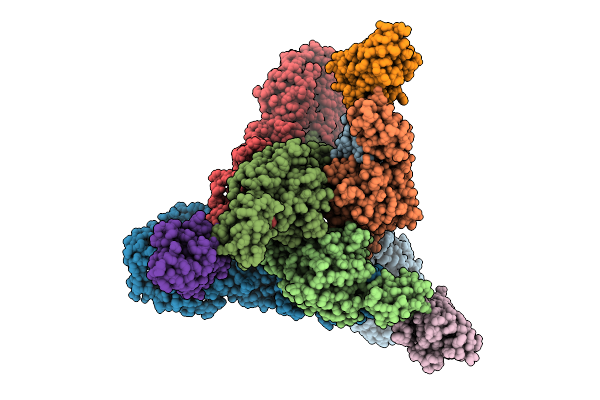
Venezuelan Equine Encephalitis Virus In Complex With The Single Domain Antibody V2C3
Organism: Venezuelan equine encephalitis virus, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: Viral protein/Immune System |
 |
Venezuelan Equine Encephalitis Virus In Complex With The Single Domain Antibody V3A8F
Organism: Venezuelan equine encephalitis virus, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: Viral protein/Immune System |
 |
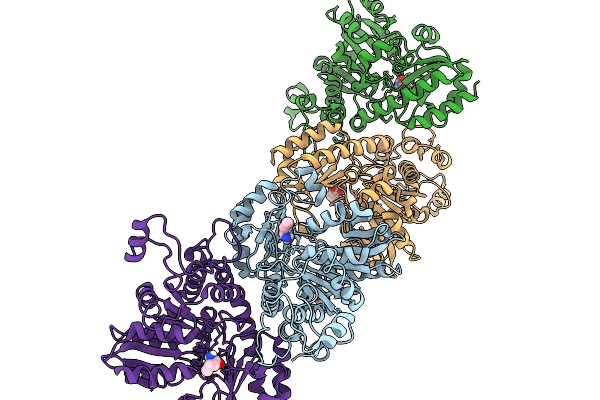
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSCRIPTION/INHIBITOR Ligands: BME, 9N6, NA, GOL, SO4 |
 |
Crystal Structure Of Highly Stable Methionine Gamma-Lyase From Thermobrachium Celere In Complex With Plp And Norleucine
Organism: Thermobrachium celere
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: LYASE Ligands: NLE, EDO |
 |
Structure Of Cathepsin B1 From Schistosoma Mansoni (Smcb1) In Complex With A Carborane Inhibitor
Organism: Schistosoma mansoni
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: A1IHQ, EDO |
 |
Crystal Structure Of Gdp-Bound Kras G12D/M67R: Suppressing G12D Oncogenicity Via Second-Site M67R Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: ONCOPROTEIN Ligands: GDP, MG |
 |
Crystal Structure Of Gdp-Bound Kras G12D/I55E: Suppressing G12D Oncogenicity Via Second-Site I55E Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: ONCOPROTEIN Ligands: GDP, MG |
 |
Crystal Structure Of Gdp-Bound Kras G12D/F28K: Suppressing G12D Oncogenicity Via Second-Site F28K Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: ONCOPROTEIN Ligands: GDP, MG, SO4 |
 |
Crystal Structure Of Gdp-Bound Kras G12D/P34R: Suppressing G12D Oncogenicity Via Second-Site P34R Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: ONCOPROTEIN Ligands: MG, GDP |
 |
Crystal Structure Of Gdp-Bound Kras G12D/R41Q: Suppressing G12D Oncogenicity Via Second-Site R41Q Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: ONCOPROTEIN Ligands: GDP, MG |
 |
Crystal Structure Of Gdp-Bound Kras G12D/V45E: Suppressing G12D Oncogenicity Via Second-Site V45E Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: ONCOPROTEIN Ligands: GDP, MG |
 |
Crystal Structure Of Gdp-Bound Kras G12D/D54R: Suppressing G12D Oncogenicity Via Second-Site D54R Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: ONCOPROTEIN Ligands: GDP, MG, SO4 |
 |
Crystal Structure Of Gdp-Bound Kras G12D/G60R: Suppressing G12D Oncogenicity Via Second-Site G60R Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: ONCOPROTEIN Ligands: MG, GDP |

