Search Count: 16
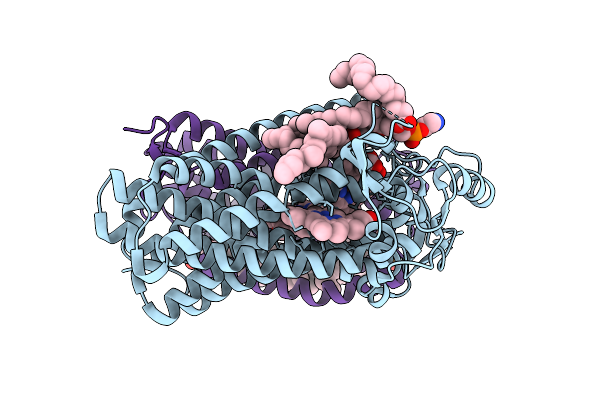 |
Cryo-Em Structure Of The Reduced Cytochrome Bd Oxidase From M. Tuberculosis
Organism: Mycobacterium tuberculosis h37rv
Method: ELECTRON MICROSCOPY Resolution:2.96 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: PTY, MQ9, HEM, HDD, CDL, CD4 |
 |
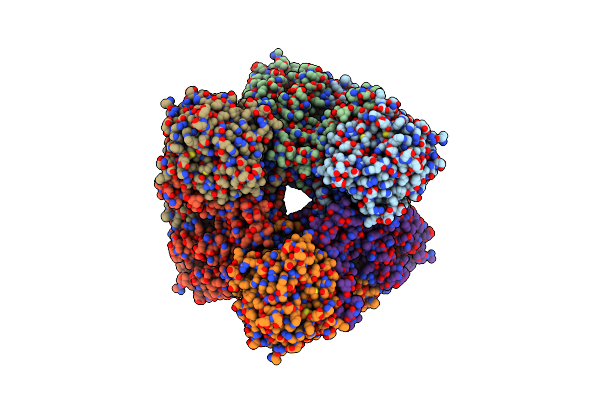
Organism: Methanothermobacter marburgensis
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: TRANSFERASE Ligands: MG, JCV, NA, COM |
 |
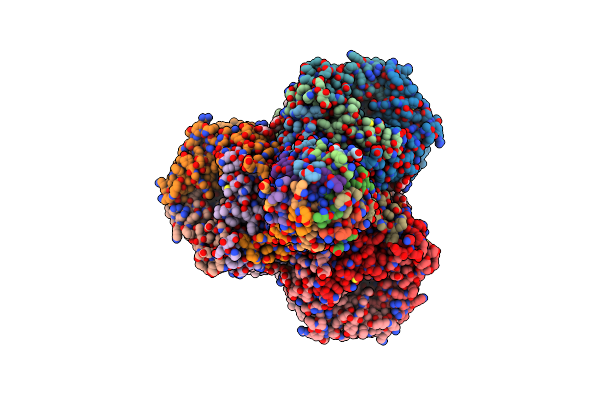
Cryo-Em Structure Of Active Phthaloyl-Coa Decarboxylase (Pcd) Complex With Prfmn Bound
Organism: Thauera chlorobenzoica
Method: ELECTRON MICROSCOPY Release Date: 2024-07-10 Classification: FLAVOPROTEIN Ligands: BYN, FE, K |
 |
Cryo-Em Structure Of The Methanogenic Na+ Translocating N5-Methyl-H4Mpt:Com Methyltransferase Complex
Organism: Methanothermobacter marburgensis
Method: ELECTRON MICROSCOPY Release Date: 2024-07-10 Classification: TRANSFERASE Ligands: MG, JCV, NA |
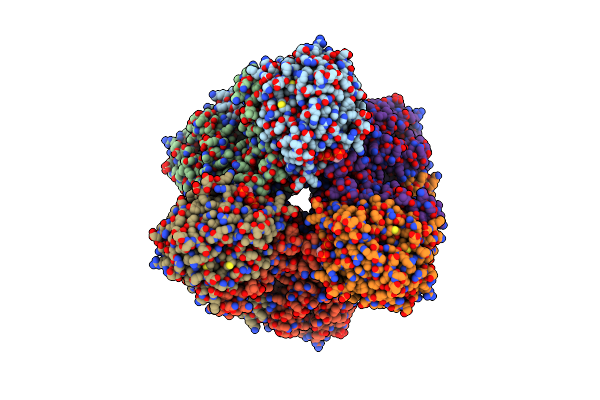 |
Cryo-Em Structure Of Phthaloyl-Coa Decarboxylase (Pcd) Bound With Substrate Analog/Inhibitor, 2-Cn-Benzoyl-Coa
Organism: Thauera chlorobenzoica
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: FLAVOPROTEIN Ligands: BYN, WC8, FE, K, CA |
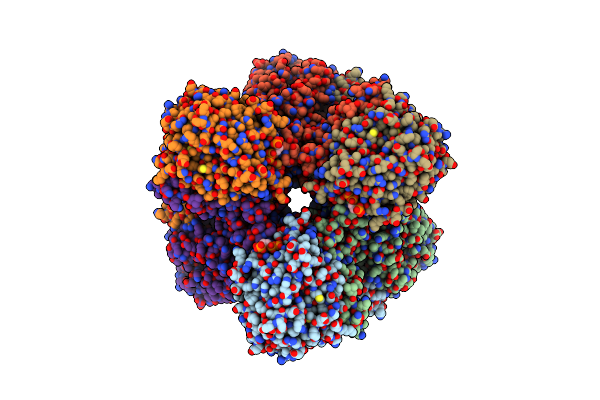 |
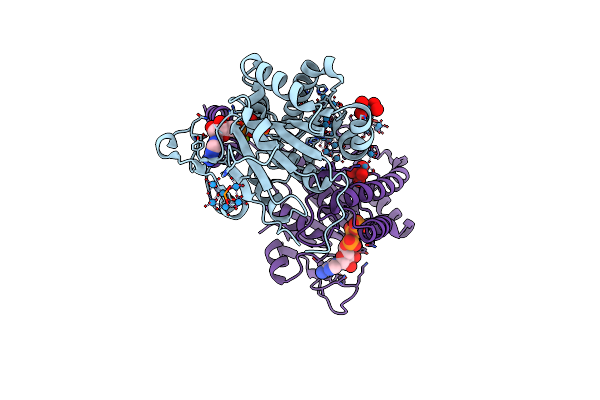
Cryo-Em Structure Of Phthaloyl-Coa Decarboxylase (Pcd) Bound With Product, Benzoyl-Coa
Organism: Thauera chlorobenzoica
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: FLAVOPROTEIN Ligands: BYC, BYN, FE, CA, K |
 |
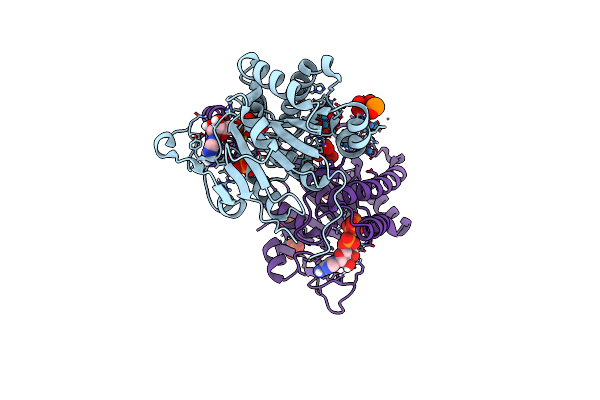
Molybdenum Storage Protein Loaded With Polyoxotungstates In The In Vivo-Like State
Organism: Azotobacter vinelandii
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-08-03 Classification: METAL BINDING PROTEIN Ligands: MG, ATP, UNL, CL |
 |
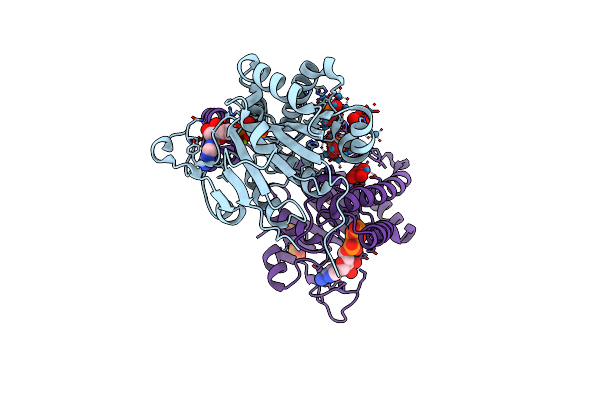
Molybdenum Storage Protein In Complex With Polyoxotungstates In The In-Vitro State
Organism: Azotobacter vinelandii
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-08-03 Classification: METAL BINDING PROTEIN Ligands: MG, ATP, UNL, PO4 |
 |
Organism: Azotobacter vinelandii
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-07-27 Classification: METAL BINDING PROTEIN Ligands: ATP, MG, UNL, CL, PO4 |
 |
Organism: Azotobacter vinelandii dj
Method: ELECTRON MICROSCOPY Release Date: 2022-07-13 Classification: METAL BINDING PROTEIN Ligands: MOO, ATP, MG, IWL, IV9, IWO, IWZ, IHW, IX3 |
 |
Organism: Acetobacterium woodii
Method: ELECTRON MICROSCOPY Release Date: 2022-06-29 Classification: FLAVOPROTEIN Ligands: FAD, FE |
 |
Organism: Hyphomicrobium denitrificans
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2020-08-19 Classification: FLAVOPROTEIN Ligands: SF4, FAD, MES |
 |
Organism: Bacterium enrichment culture clone n47
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-05-15 Classification: FLAVOPROTEIN Ligands: SF4, FAD, FMN |
 |
Organism: Bacterium enrichment culture clone n47
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-05-15 Classification: FLAVOPROTEIN Ligands: SF4, FAD, FMN, J5H |
 |
2-Naphthoyl-Coa Reductase-Dihydronaphthoyl-Coa Complex(Ncr-Dhncoa Co-Crystallized Complex)
Organism: Bacterium enrichment culture clone n47
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-05-15 Classification: FLAVOPROTEIN Ligands: SF4, FAD, FMN, J5H |
 |
Molecular Basis Of The Flavin-Based Electron-Bifurcating Caffeyl-Coa Reductase Reaction
Organism: Acetobacterium woodii (strain atcc 29683 / dsm 1030 / jcm 2381 / kctc 1655 / wb1)
Method: X-RAY DIFFRACTION Release Date: 2018-01-24 Classification: FLAVOPROTEIN Ligands: FAD, SF4, SO4 |

