Search Count: 22
 |
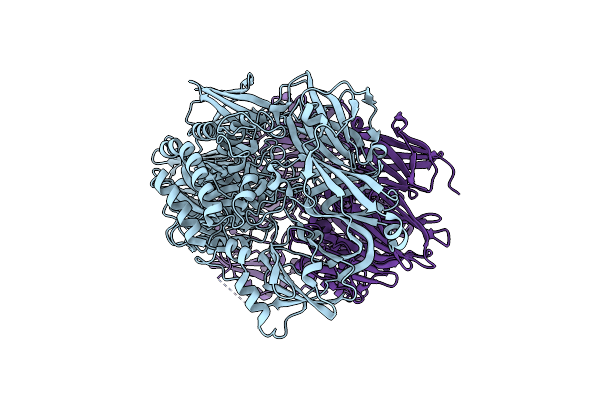
Organism: Tick-borne encephalitis virus (strain sofjin)
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2024-10-09 Classification: VIRAL PROTEIN |
 |
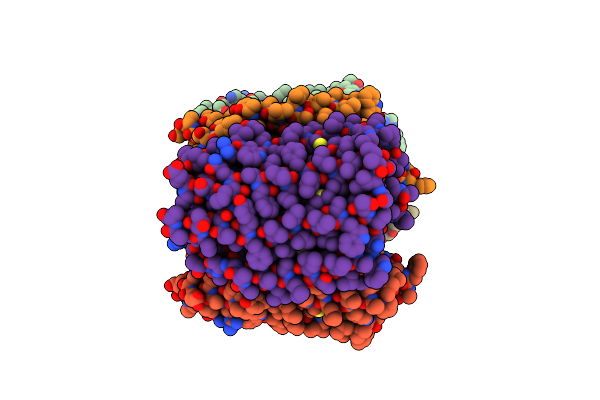
Organism: Desulfurococcus amylolyticus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2024-08-14 Classification: HYDROLASE |
 |
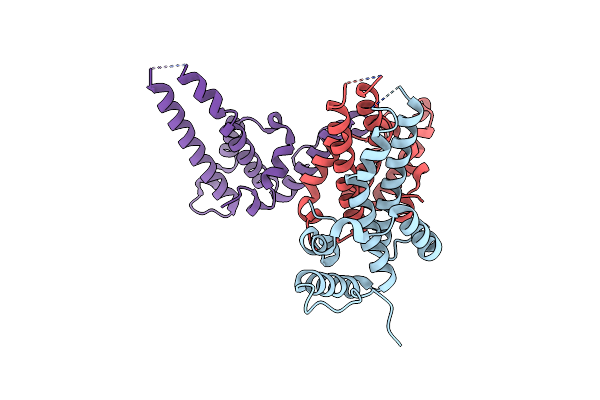
Crystal Structure Of The Microbial Rhodopsin From Sphingomonas Paucimobilis (Spar)
Organism: Sphingomonas paucimobilis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-05-03 Classification: MEMBRANE PROTEIN Ligands: LFA |
 |
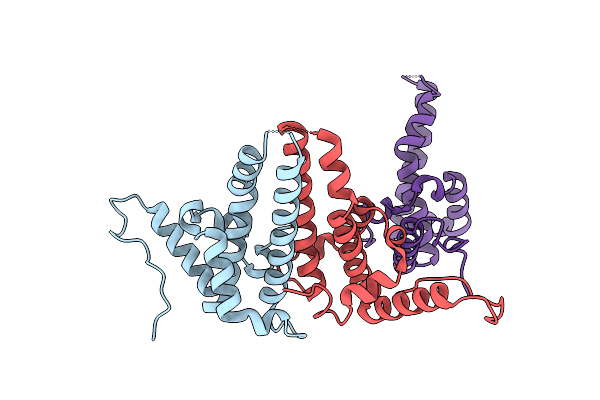
Hepatitis B Virus Core Antigen (Hbc) With The Insertion Of Four External Domains Of The Influenza A M2 Protein (Hbc/4M2E) With T=4 Topology
Organism: Hepatitis b virus adw/991, Influenza a virus (a/malaya/302/1954(h1n1)), Hepatitis b virus genotype d subtype ayw (isolate france/tiollais/1979)
Method: ELECTRON MICROSCOPY Release Date: 2022-12-28 Classification: VIRUS LIKE PARTICLE |
 |
Hepatitis B Virus Core Antigen (Hbc) With The Insertion Of Four External Domains Of The Influenza A M2 Protein (Hbc/4M2E) With T=3 Topology
Organism: Hepatitis b virus, Influenza a virus (strain a/malaysia:malaya/302/1954 h1n1)
Method: ELECTRON MICROSCOPY Release Date: 2022-12-28 Classification: VIRUS LIKE PARTICLE |
 |
Dynamic Complex Between All-D-Enantiomeric Peptide D3 With Wild-Type Amyloid Precursor Protein 672-726 Fragment (Amyloid Beta 1-55)
Organism: Homo sapiens, Synthetic construct
Method: SOLUTION NMR Release Date: 2021-01-13 Classification: PROTEIN BINDING |
 |
Dynamic Complex Between All-D-Enantiomeric Peptide D3 With L723P Mutant Of Amyloid Precursor Protein (App) 672-726 Fragment (Amyloid Beta 1-55)
Organism: Homo sapiens, Synthetic construct
Method: SOLUTION NMR Release Date: 2021-01-13 Classification: PROTEIN BINDING |
 |
Crystal Structure Of Hepatitis C Virus Ires Junction Iiiabc In Complex With Fab Hcv2
Organism: Homo sapiens, Hepacivirus c
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2019-12-04 Classification: RNA/Immune System |
 |
Crystal Structure Of Hepatitis C Virus Ires Junction Iiiabc In Complex With Fab Hcv3
Organism: Homo sapiens, Hepacivirus c
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2019-12-04 Classification: RNA/Immune System |
 |
Crystal Structure Of Hepatitis A Virus Ires Domain V In Complex With Fab Havx
Organism: Homo sapiens, Hepatovirus a
Method: X-RAY DIFFRACTION Resolution:2.84 Å Release Date: 2019-08-14 Classification: RNA/Immune System |
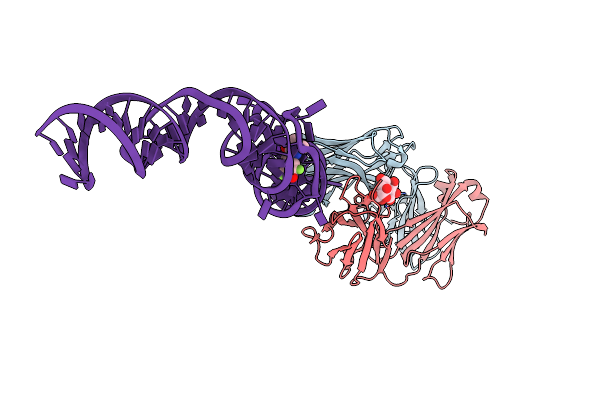 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2014-06-18 Classification: immune system/rna Ligands: K, MG, 1TU |
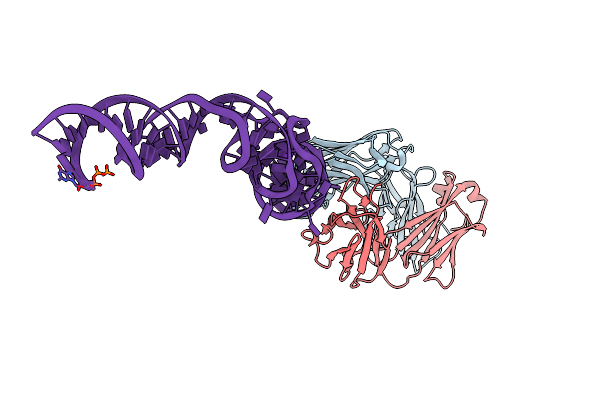 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-06-18 Classification: immune system/rna Ligands: K, MG |
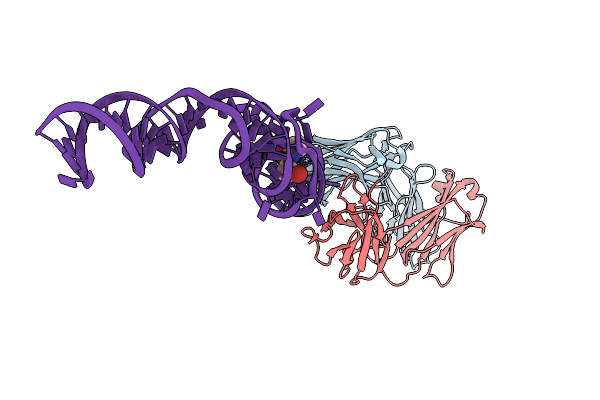 |
Crystal Structure Of An Rna Aptamer Bound To Bromo-Ligand Analog In Complex With Fab
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2014-06-18 Classification: RNA Ligands: MG, K, 2ZZ |
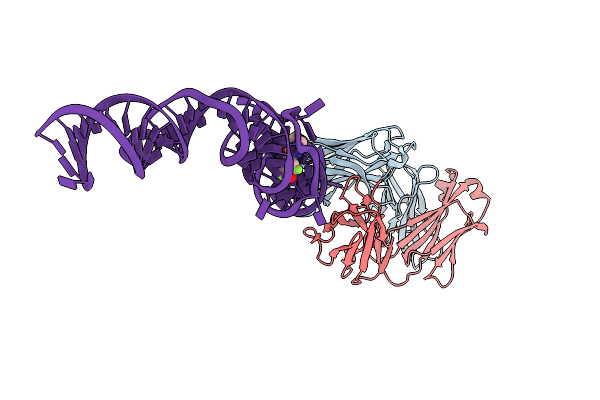 |
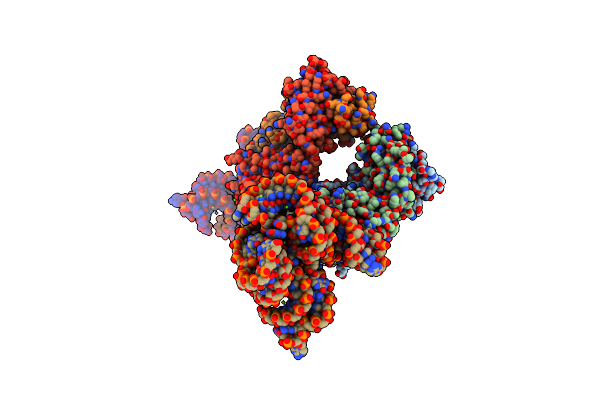
Crystal Structure Of An Rna Aptamer Bound To Trifluoroethyl-Ligand Analog In Complex With Fab
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.12 Å Release Date: 2014-06-18 Classification: RNA/immune system Ligands: K, 2ZY |
 |
Crystal Structure Of The Catalytic Core Of An Rna Polymerase Ribozyme Complexed With An Antigen Binding Antibody Fragment
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2010-03-02 Classification: immune system / RNA Ligands: CD, MG, CL |
 |
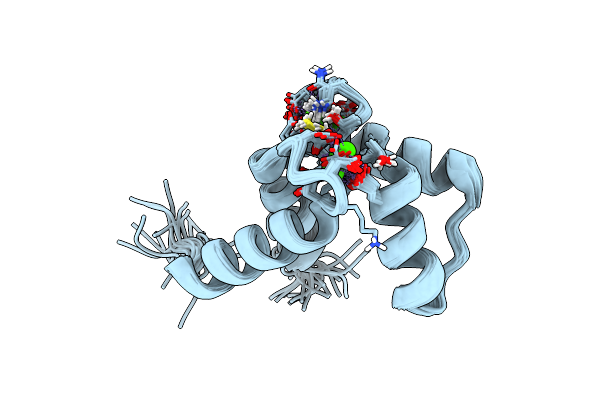
Crystal Structure Of Class I Ligase Ribozyme Self-Ligation Product, In Complex With U1A Rbd
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2009-11-24 Classification: LIGASE/RNA Ligands: MG |
 |
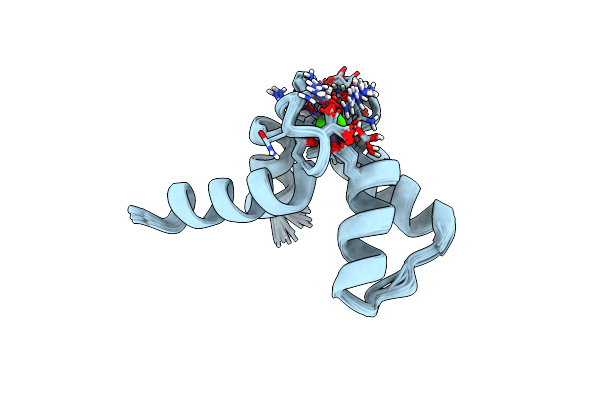
Solution Structure Of Calcium Bound Soybean Calmodulin Isoform 1 N-Terminal Domain
Organism: Glycine max
Method: SOLUTION NMR Release Date: 2008-04-08 Classification: METAL BINDING PROTEIN Ligands: CA |
 |
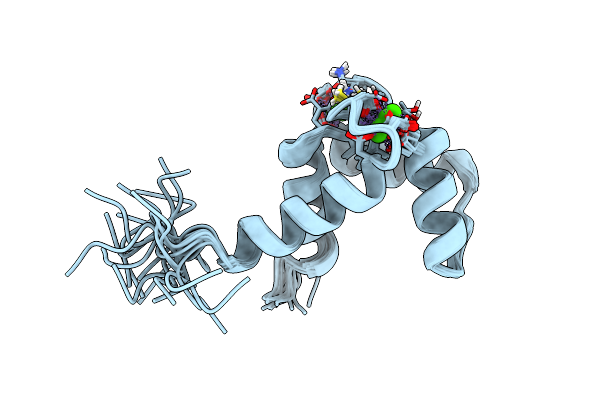
Solution Structure Of Calcium Bound Soybean Calmodulin Isoform 1 C-Terminal Domain
Organism: Glycine max
Method: SOLUTION NMR Release Date: 2008-04-08 Classification: METAL BINDING PROTEIN Ligands: CA |
 |
Solution Structure Of Calcium Bound Soybean Calmodulin Isoform 4 N-Terminal Domain
Organism: Glycine max
Method: SOLUTION NMR Release Date: 2008-04-08 Classification: METAL BINDING PROTEIN Ligands: CA |
 |
Solution Structure Of Calcium Bound Soybean Calmodulin Isoform 4 C-Terminal Domain
Organism: Glycine max
Method: SOLUTION NMR Release Date: 2008-04-08 Classification: METAL BINDING PROTEIN Ligands: CA |

