Search Count: 35
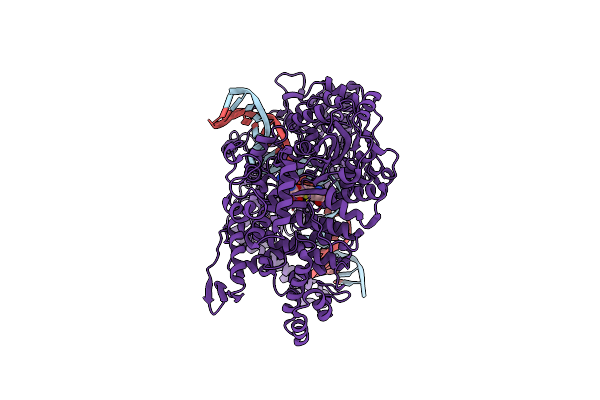 |
Cryo-Em Structure Of The E. Coli Brxx Methyltransferase In Complex With Dna
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: TRANSFERASE Ligands: MG, SAM |
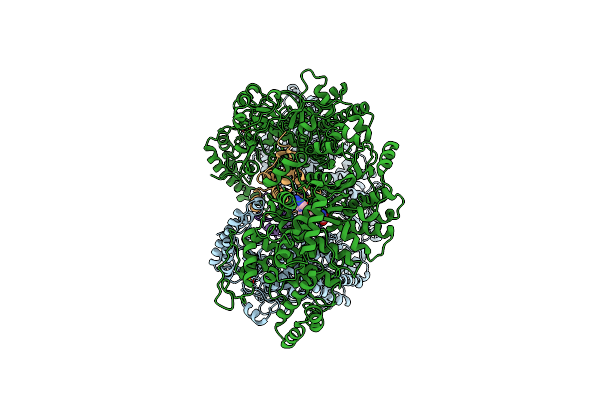 |
Cryo-Em Structure Of The E. Coli Brxx Methyltransferase In Complex With Ocr
Organism: Escherichia coli, Escherichia phage t7
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: TRANSFERASE Ligands: MG, SAM |
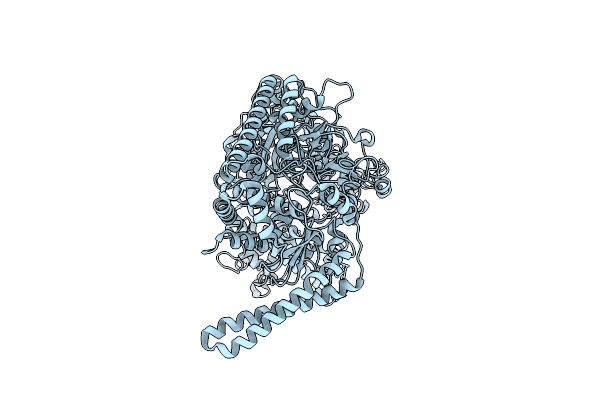 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: TRANSFERASE |
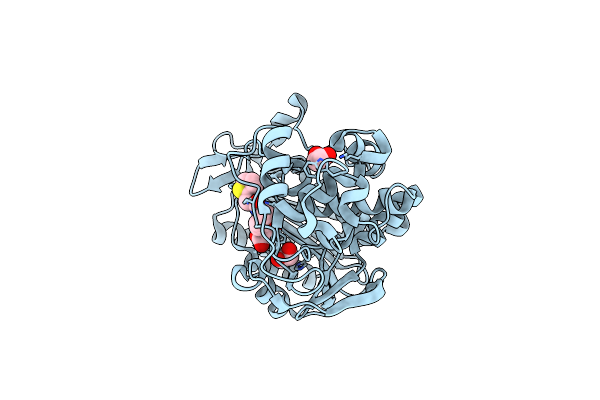 |
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-01-25 Classification: HYDROLASE Ligands: ZXQ, EDO, CL |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-10-12 Classification: HYDROLASE Ligands: ZXQ, EDO |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-05-18 Classification: HYDROLASE Ligands: ZXQ, SO4, EDO, CL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2020-08-12 Classification: HYDROLASE Ligands: 4D6, WXM, PO4, K |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2020-08-12 Classification: HYDROLASE Ligands: 4D6, SO4, GOL |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-03-25 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: RM9, QNA, GOL, SO4 |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2020-03-25 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: ZN, QNA |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-03-25 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: RM9, MG, CL |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2020-03-25 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: QNA, ZN, ACT, GOL |
 |
Structure Of A Functional Obligate Respiratory Supercomplex From Mycobacterium Smegmatis
Organism: Mycobacterium smegmatis mc2 155, Mycobacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: ELECTRON MICROSCOPY Release Date: 2018-11-07 Classification: ELECTRON TRANSPORT Ligands: FES, CDL, MQ9, CU, HAS, HEC, HEM |
 |
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2015-04-01 Classification: Hydrolase/antibiotic Ligands: 4D6, EDO, PG0 |
 |
Organism: Klebsiella pneumoniae subsp. pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-04-01 Classification: Hydrolase/antibiotic Ligands: CL, CIT, EDO, 4D6 |
 |
Crystal Structure Of Runx1 And Ets1 Bound To Tcr Alpha Promoter (Crystal Form 1)
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-03-26 Classification: TRANSCRIPTION/DNA |
 |
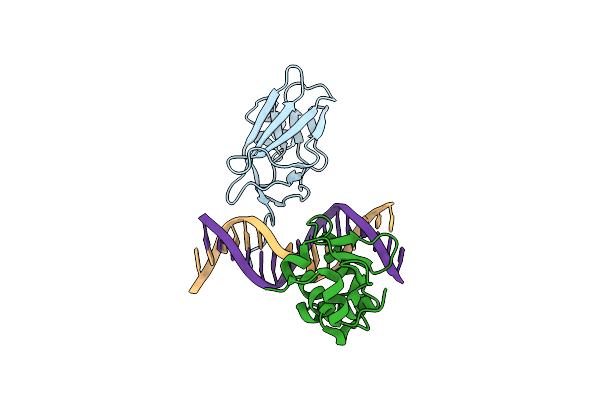
Crystal Structure Of Runx1 And Ets1 Bound To Tcr Alpha Promoter (Crystal Form 2)
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2014-03-26 Classification: TRANSCRIPTION/DNA |
 |
Crystal Structure Of Runx1 And Ets1 Bound To Tcr Alpha Promoter (Crystal Form 3)
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-03-26 Classification: TRANSCRIPTION/DNA Ligands: GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2012-04-04 Classification: Transcription/DNA |
 |
Crystal Structure Of The 2'- Deoxyguanosine Riboswitch Bound To 2'-Deoxyguanosine
Method: X-RAY DIFFRACTION
Resolution:2.30 Å Release Date: 2011-08-17 Classification: RNA Ligands: GNG, MG, SIN, SO4 |

