Search Count: 24
 |
Crystal Structure Of Aedes Aegypti Noppera-Bo, Glutathione S-Transferase Epsilon 8, In Glutathione-Bound Form
Organism: Aedes aegypti
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2022-01-26 Classification: TRANSFERASE Ligands: GSH, CA |
 |
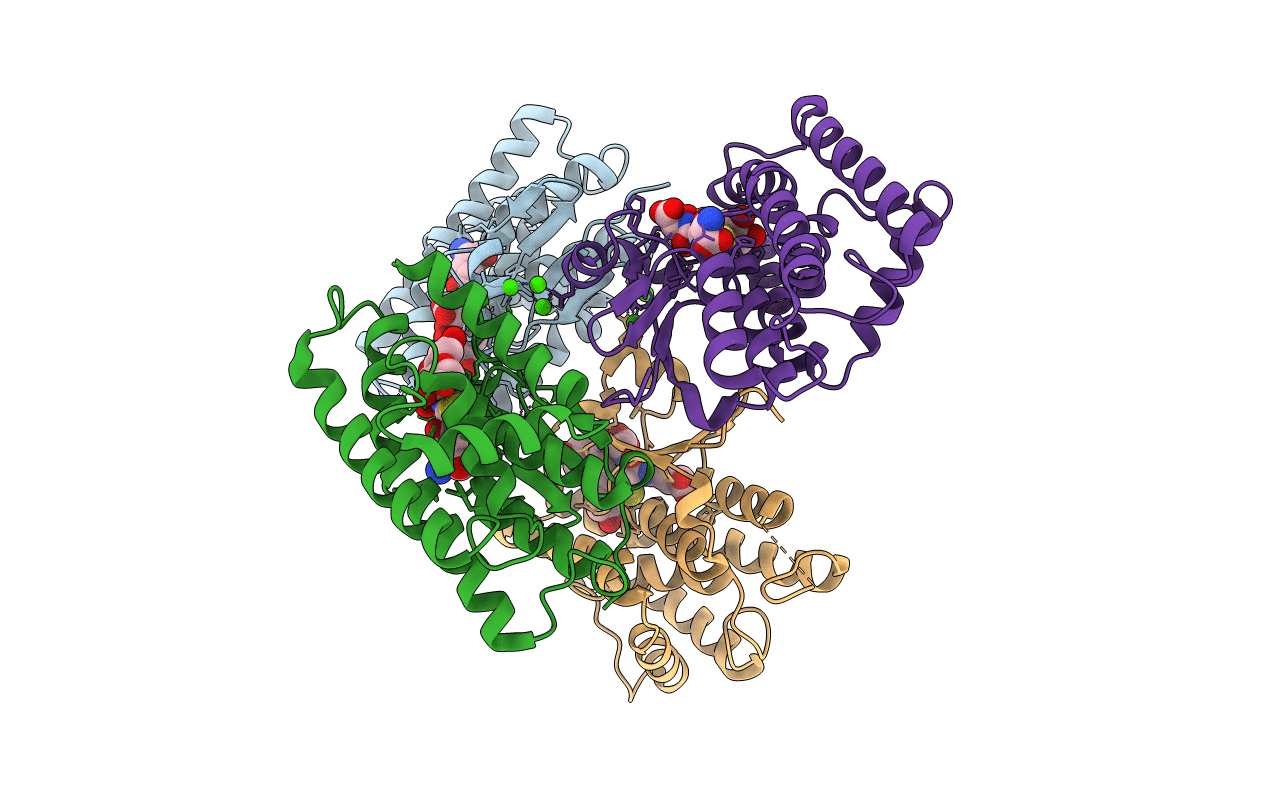
Crystal Structure Of Aedes Aegypti Noppera-Bo, Glutathione S-Transferase Epsilon 8, In Daidzein- And Glutathione-Bound Form
Organism: Aedes aegypti
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-01-26 Classification: TRANSFERASE Ligands: GSH, ZF1, CA |
 |
Crystal Structure Of Aedes Aegypti Noppera-Bo, Glutathione S-Transferase Epsilon 8, In Luteolin- And Glutathione-Bound Form
Organism: Aedes aegypti
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-01-26 Classification: TRANSFERASE Ligands: GSH, LU2, CA |
 |
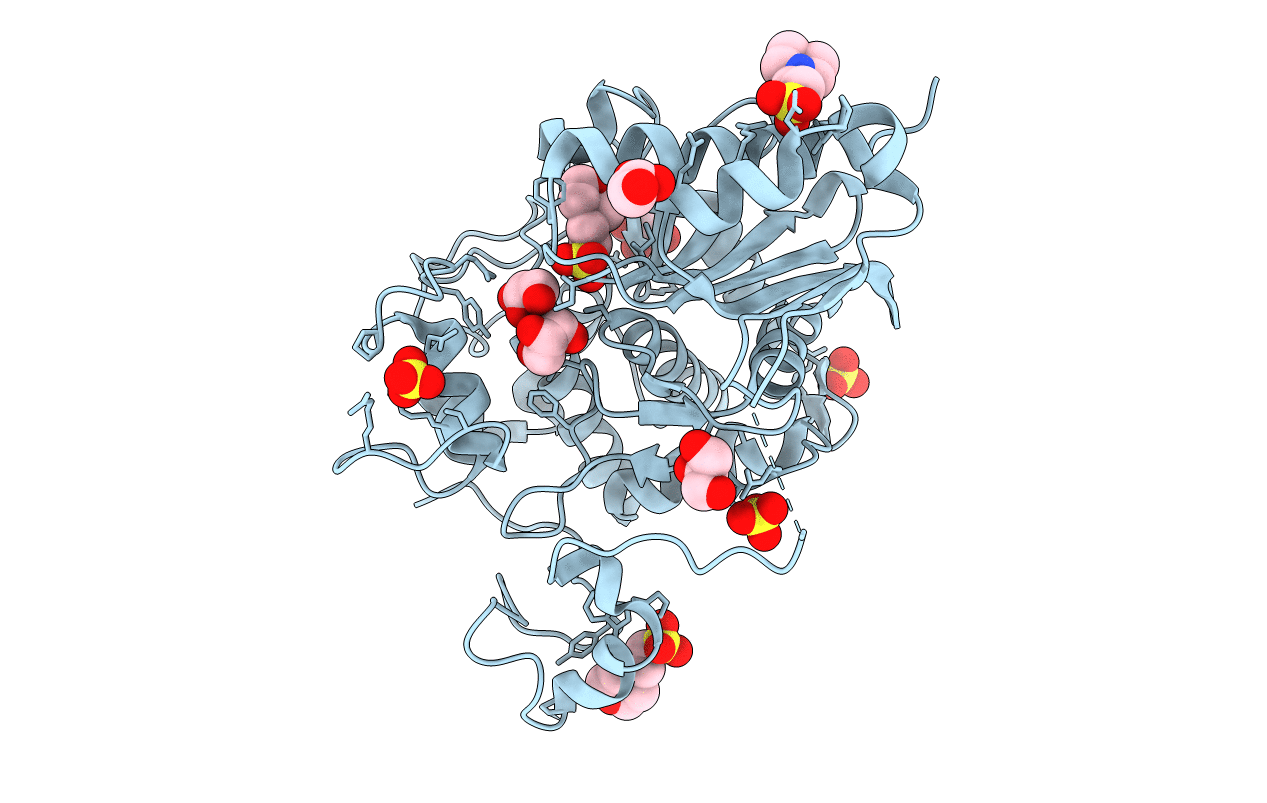
Crystal Structure Of Aedes Aegypti Noppera-Bo, Glutathione S-Transferase Epsilon 8, In Desmethylglycitein And Glutathione-Bound Form
Organism: Aedes aegypti
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2022-01-26 Classification: TRANSFERASE Ligands: GSH, 674, CA |
 |
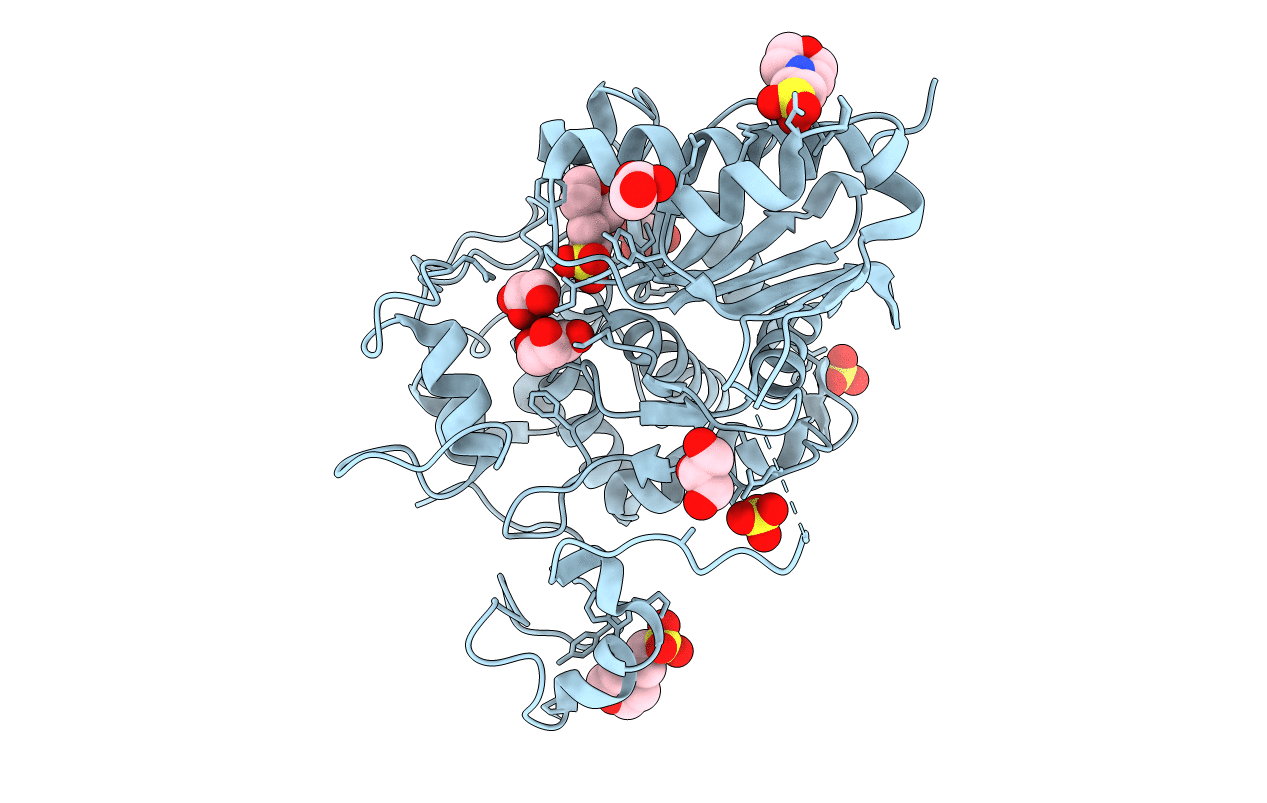
Crystal Structure Of 6-Aminohexanoate-Dimer Hydrolase G181D/R187S/H266N/D370Y Mutant
Organism: Flavobacterium
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-10-16 Classification: HYDROLASE Ligands: GOL, MES, SO4 |
 |
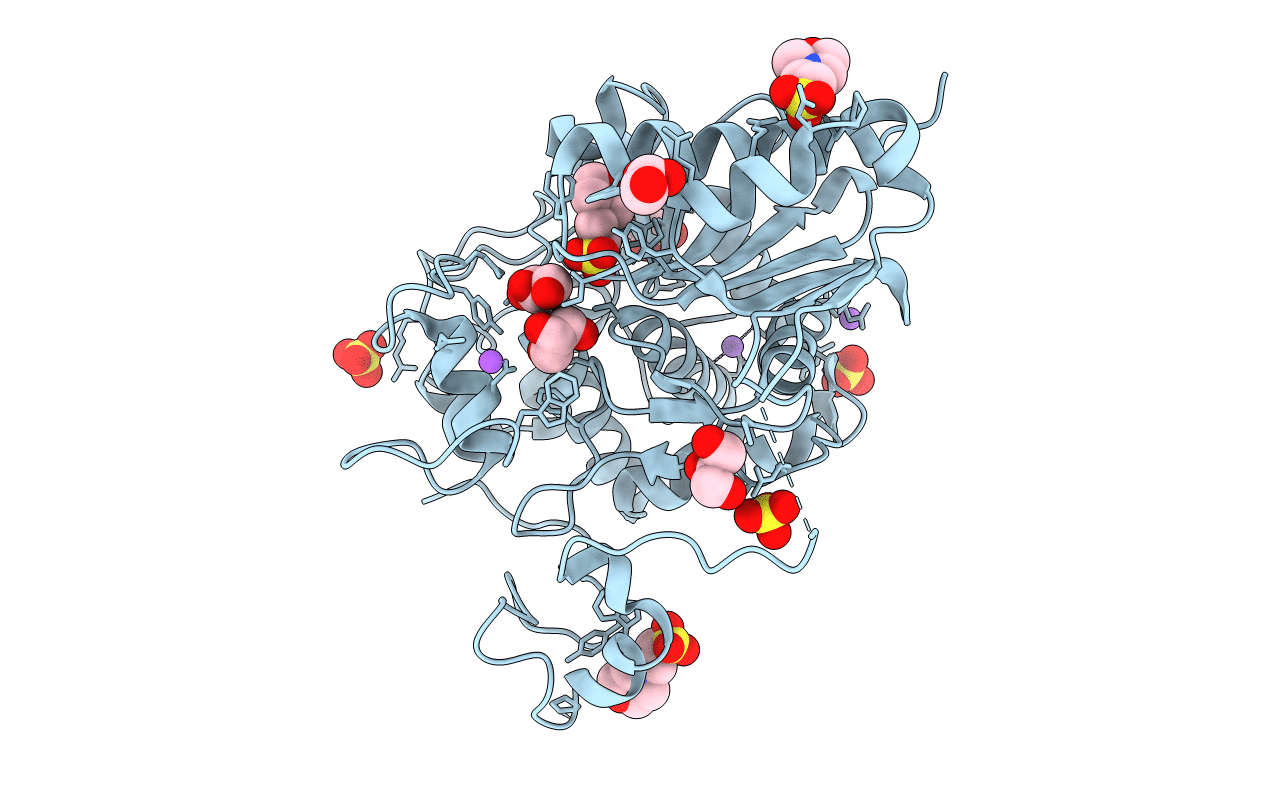
Crystal Structure Of 6-Aminohexanoate-Dimer Hydrolase G181D/R187A/H266N/D370Y Mutant
Organism: Flavobacterium
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-10-16 Classification: HYDROLASE Ligands: GOL, MES, SO4 |
 |
Crystal Structure Of 6-Aminohexanoate-Dimer Hydrolase G181D/R187G/H266N/D370Y Mutant
Organism: Flavobacterium
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2013-10-16 Classification: HYDROLASE Ligands: GOL, MES, SO4, NA |
 |
Crystal Structure Of 6-Aminohexanoate-Dimer Hydrolase S112A/G181D/R187S/H266N/D370Y Mutant Complexd With 6-Aminohexanoate
Organism: Flavobacterium
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2013-10-16 Classification: HYDROLASE |
 |
6-Aminohexanoate-Dimer Hydrolase S112A/G181D/R187A/H266N/D370Y Mutant Complexd With 6-Aminohexanoate
Organism: Flavobacterium
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-10-16 Classification: HYDROLASE Ligands: ACA, GOL, MES |
 |
Crystal Structure Of 6-Aminohexanoate-Dimer Hydrolase S112A/G181D/R187G/H266N/D370Y Mutant Complexd With 6-Aminohexanoate
Organism: Flavobacterium
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2013-10-16 Classification: HYDROLASE Ligands: ACA, GOL, MES, SO4 |
 |
Crystal Structure Of Hla-B*5201 In Complexed With Hiv Immunodominant Epitope (Taftipsi)
Organism: Homo sapiens, Human immunodeficiency virus type 1 (z2/cdc-z34 isolate)
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2013-02-13 Classification: IMMUNE SYSTEM |
 |
Crystal Structure Of 6-Aminohexanoate-Dimer Hydrolase S112A/G181D/H266N Mutant With Substrate
Organism: Flavobacterium
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2010-09-01 Classification: HYDROLASE Ligands: ACA, MES, GOL, NA |
 |
Crystal Structure Of 6-Aminohexanoate-Dimer Hydrolase S112A/G181D/H266N/D370Y Mutant With Substrate
Organism: Flavobacterium
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-09-01 Classification: HYDROLASE |
 |
Organism: Arthrobacter sp.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2009-11-03 Classification: HYDROLASE Ligands: GOL |
 |
Structure Of 6-Aminohexanoate Cyclic Dimer Hydrolase Complexed With Substrate
Organism: Arthrobacter sp.
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-11-03 Classification: HYDROLASE Ligands: ACA, GOL |
 |
Organism: Flavobacterium sp.
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2009-04-21 Classification: HYDROLASE Ligands: SO4, MES, GOL |
 |
Structure Of 6-Aminohexanoate-Dimer Hydrolase, A61V/A124V/R187S/F264C/G291R/G338A/D370Y Mutant (Hyb-S4M94)
Organism: Flavobacterium sp.
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2009-04-21 Classification: HYDROLASE Ligands: SO4, MES, GOL |
 |
Structure Of 6-Aminohexanoate-Dimer Hydrolase, S112A/D370Y Mutant Complexed With 6-Aminohexanoate-Dimer
Organism: Flavobacterium sp.
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2009-04-14 Classification: HYDROLASE Ligands: SO4, ACA, MES, GOL |
 |
Structure Of 6-Aminohexanoate-Dimer Hydrolase, A61V/S112A/A124V/R187S/F264C/G291R/G338A/D370Y Mutant (Hyb-S4M94) With Substrate
Organism: Flavobacterium sp.
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2009-04-14 Classification: HYDROLASE Ligands: SO4, ACA, MES, GOL |
 |
Organism: Flavobacterium sp.
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2009-04-07 Classification: HYDROLASE Ligands: SO4, MES, GOL |

