Search Count: 15
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-04-03 Classification: FLUORESCENT PROTEIN |
 |
Organism: Thermococcus kodakarensis (strain atcc baa-918 / jcm 12380 / kod1)
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2018-06-20 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
Organism: Thermococcus kodakarensis (strain atcc baa-918 / jcm 12380 / kod1)
Method: X-RAY DIFFRACTION Resolution:3.24 Å Release Date: 2018-06-20 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-06-22 Classification: HYDROLASE ACTIVATOR/SIGNALING PROTEIN Ligands: MG, GDP, AF3 |
 |
Crystal Structure Of The Fusion Protein Linked By Rhoa And The Gap Domain Of Mgcracgap
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2016-06-22 Classification: HYDROLASE ACTIVATOR Ligands: MG, AF3, GDP |
 |
Organism: Thermococcus kodakaraensis (strain atcc baa-918 / jcm 12380 / kod1)
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2015-06-24 Classification: METAL BINDING PROTEIN/HYDROLASE Ligands: ZN, NI, URE, CL, GOL, ADP, MG |
 |
Organism: Thermococcus kodakaraensis (strain atcc baa-918 / jcm 12380 / kod1)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-06-24 Classification: METAL BINDING PROTEIN/HYDROLASE Ligands: ZN, NI, CL, ACP, MG, GOL |
 |
Organism: Thermococcus kodakaraensis (strain atcc baa-918 / jcm 12380 / kod1)
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2015-06-24 Classification: METAL BINDING PROTEIN/HYDROLASE Ligands: ZN, ACP, MG |
 |
Organism: Thermococcus kodakaraensis (strain atcc baa-918 / jcm 12380 / kod1)
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2015-06-24 Classification: HYDROLASE Ligands: GOL |
 |
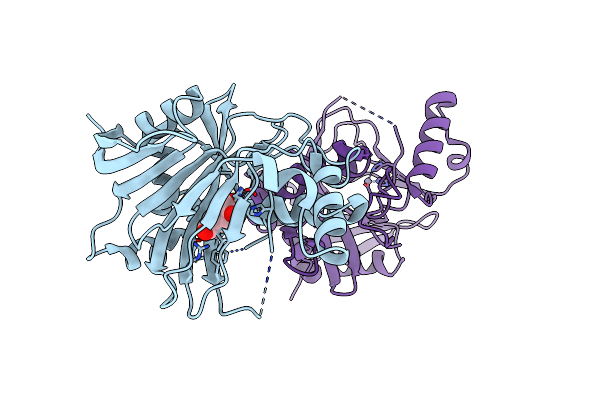
Crystal Structure Of A Novel N-Substituted L-Amino Acid Dioxygenase From Burkholderia Ambifaria Ammd
Organism: Burkholderia ambifaria
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2013-07-17 Classification: OXIDOREDUCTASE Ligands: ZN |
 |
Crystal Structure Of A Novel N-Substituted L-Amino Acid Dioxygenase In Complex With Alpha-Kg From Burkholderia Ambifaria Ammd
Organism: Burkholderia ambifaria
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2013-07-17 Classification: OXIDOREDUCTASE Ligands: ZN, AKG |
 |
Structure Of Putative Hth-Type Transcriptional Regulator Ph1519/Dna Complex
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2007-12-04 Classification: TRANSCRIPTION/DNA |
 |
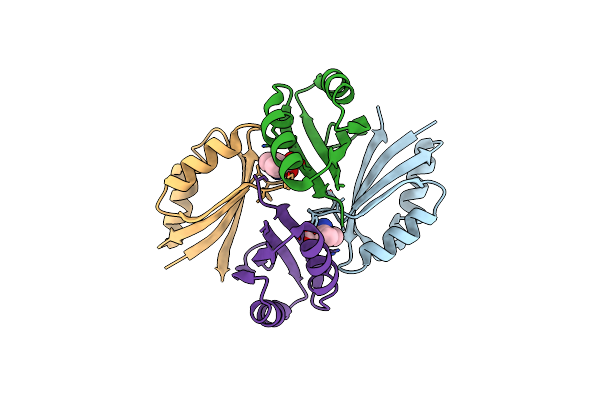
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-09-18 Classification: TRANSCRIPTION Ligands: MSE |
 |
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2007-09-18 Classification: DNA BINDING PROTEIN Ligands: ILE |
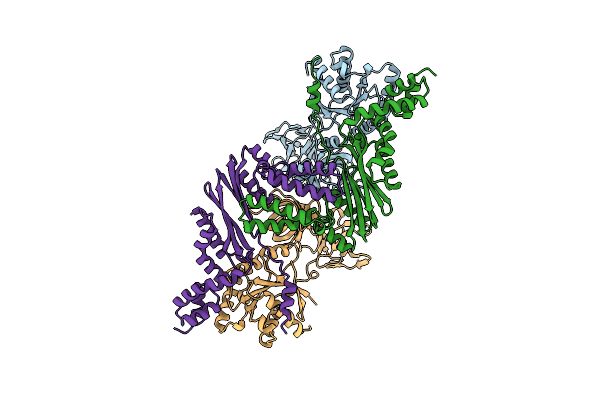 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 1997-01-11 Classification: COMPLEX (TWO ELONGATION FACTORS) |

