Search Count: 16
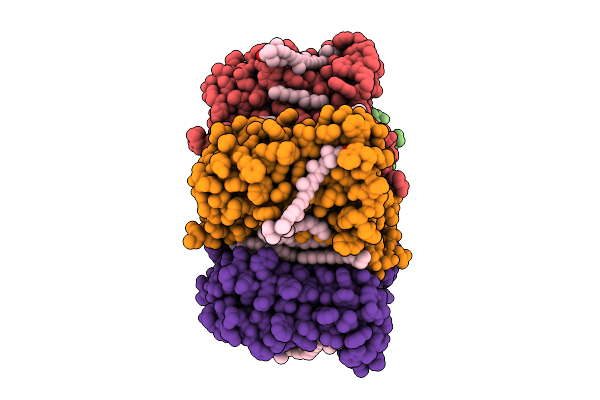 |
Cryo-Em Structure Of The Zeaxanthin-Bound Light-Driven Proton Pumping Rhodopsin, Nm-R1
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, R16, D12, K3I |
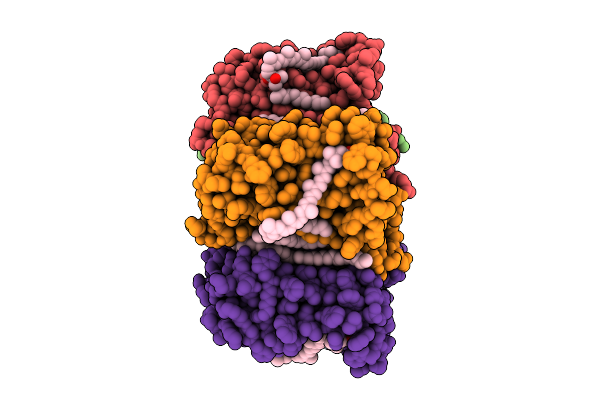 |
Cryo-Em Structure Of The Myxol-Bound Light-Driven Proton Pumping Rhodopsin, Nm-R1
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, R16, D12, A1L4O |
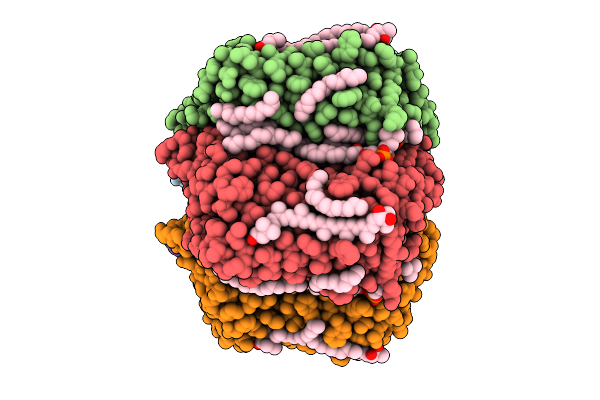 |
Cryo-Em Structure Of The Myxol-Bound Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, A1L4O, CL, PC1, PLC, R16, 8K6, D12, C14, D10 |
 |
Cryo-Em Structure Of The Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, CL, PC1, PLC, D12, R16, 8K6, C14 |
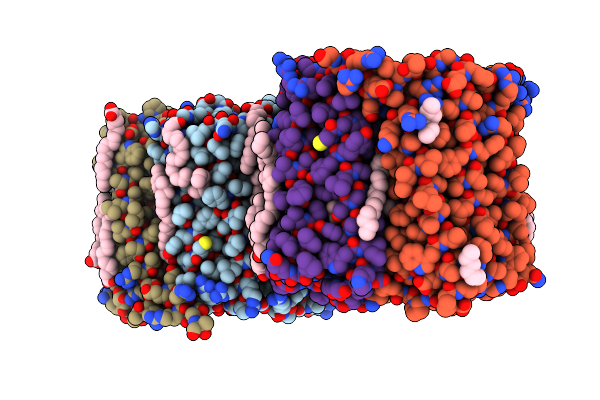 |
Organism: Salinarimonas soli
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2024-10-16 Classification: MEMBRANE PROTEIN Ligands: LFA, CL, OLA, RET |
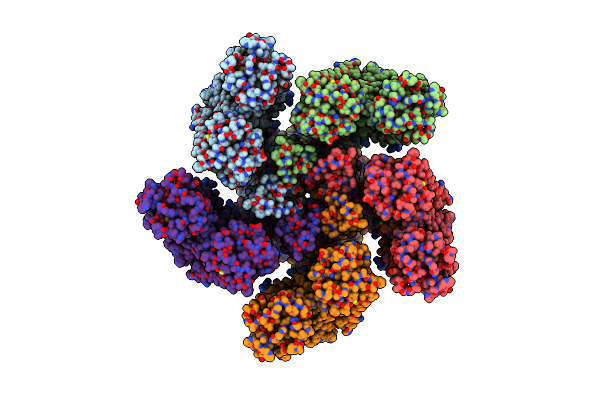 |
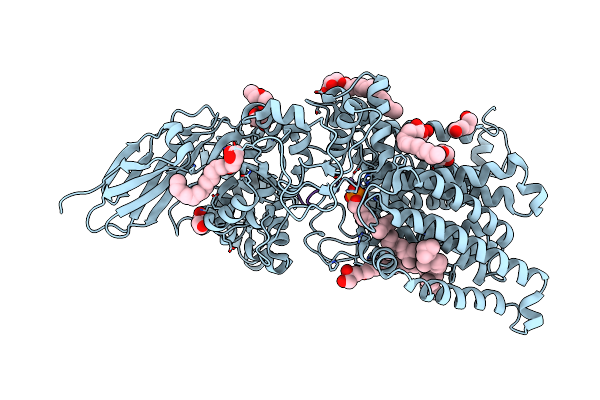
Cryo-Em Structure Of Bestrhodopsin (Rhodopsin-Rhodopsin-Bestrophin) Complex
Organism: Phaeocystis
Method: ELECTRON MICROSCOPY Release Date: 2022-07-06 Classification: MEMBRANE PROTEIN Ligands: RET |
 |
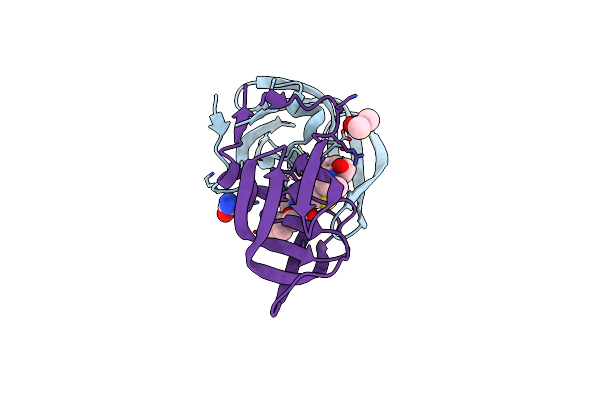
Archaeal Oligosaccharyltransferase Aglb From Archaeoglobus Fulgidus In Complex With An Inhibitory Peptide And A Dolichol-Phosphate
Organism: Archaeoglobus fulgidus dsm 4304, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2021-09-08 Classification: TRANSFERASE Ligands: MN, J06, PEG, 7E8 |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-09-07 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: GOL, URE, 016, DMS |
 |
Crystal Structure Of Hiv-1 Protease (Q7K, L33I, L63I) In Complex With Kni-10074
Organism: Human immunodeficiency virus type 1
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-03-16 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: JZP, GOL, CL |
 |
Crystal Structure Of Hiv-1 Protease (Q7K, L33I, L63I) In Complex With Kni-10006
Organism: Human immunodeficiency virus type 1
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2010-03-02 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: GOL, 006 |
 |
Crystal Structure Of Hiv-1 Protease (Q7K, L33I, L63I) In Complex With Kni-10265
Organism: Human immunodeficiency virus type 1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-03-02 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: JZQ, GOL |
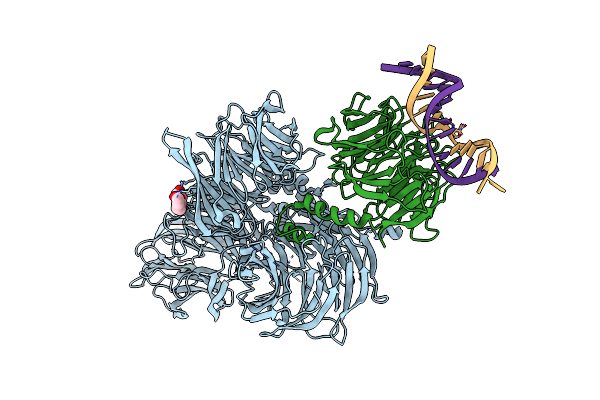 |
Structure Of Hsddb1-Drddb2 Bound To A 14 Bp 6-4 Photoproduct Containing Dna-Duplex
Organism: Homo sapiens, Danio rerio
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2009-01-20 Classification: DNA BINDING PROTEIN/DNA Ligands: PG4 |
 |
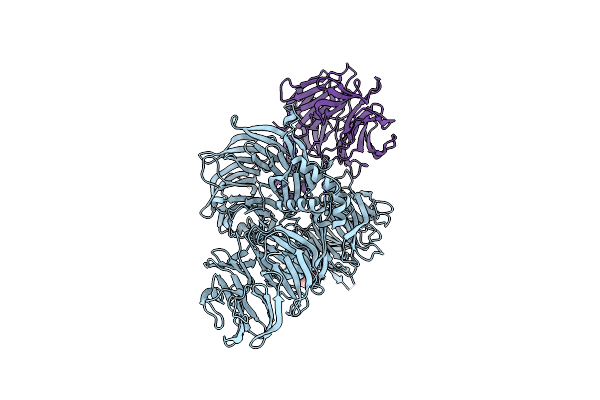
Structure Of Hsddb1-Drddb2 Bound To A 16 Bp Abasic Site Containing Dna-Duplex
Organism: Homo sapiens, Danio rerio
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2009-01-20 Classification: DNA BINDING PROTEIN/DNA Ligands: PG4 |
 |
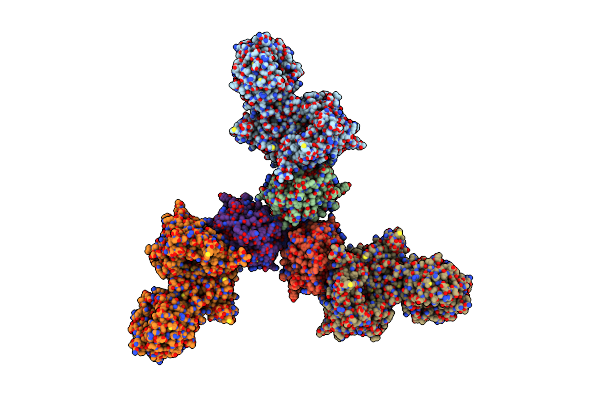
Organism: Homo sapiens, Danio rerio
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2009-01-20 Classification: DNA BINDING PROTEIN Ligands: PG4 |
 |
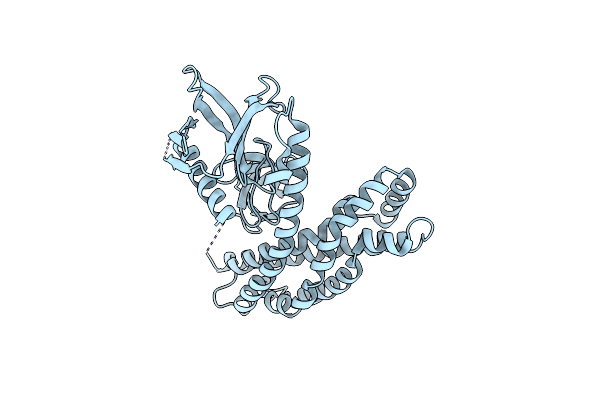
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2009-01-20 Classification: DNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2007-01-02 Classification: SIGNALING PROTEIN |

