Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Kutzneria albida dsm 43870
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: LIGASE Ligands: MG, AMP |
 |
Organism: Kutzneria albida dsm 43870
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: LIGASE |
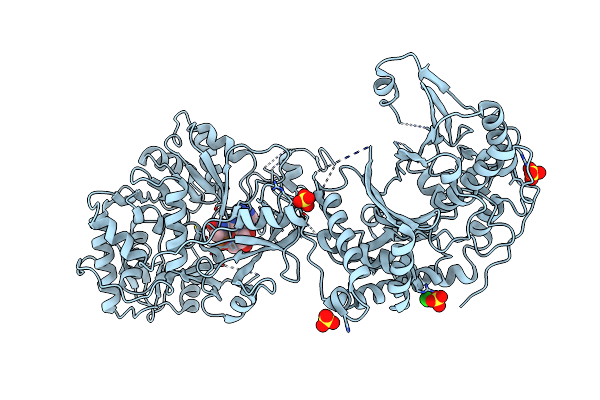 |
Crystal Structure Of Nonribosomal Peptide Synthetases (Nrps), Fmoa3 (S1046A)-Alpha-Methyl-L-Serine-Amp Bound Form
Organism: Streptomyces sp. sp080513ge-23
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2021-03-10 Classification: BIOSYNTHETIC PROTEIN Ligands: AMP, EW6, SO4, CL |
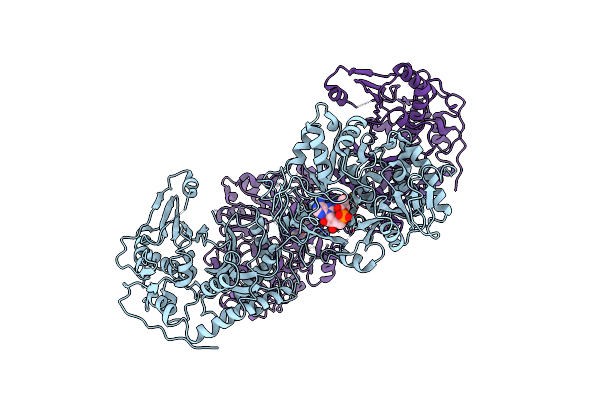 |
Crystal Structure Of Nonribosomal Peptide Synthetases (Nrps), Fmoa3 (S1046A)-Alpha-Methyl-L-Serine-Amp Bound Form
Organism: Streptomyces sp. sp080513ge-23
Method: X-RAY DIFFRACTION Resolution:4.10 Å Release Date: 2021-03-10 Classification: BIOSYNTHETIC PROTEIN Ligands: AMP, EW6 |
 |
Crystal Structure Of Nonribosomal Peptide Synthetases (Nrps), Fmoa3 (S1046A)
Organism: Streptomyces sp. sp080513ge-23
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2021-02-24 Classification: BIOSYNTHETIC PROTEIN Ligands: AKR |
 |
Crystal Structure Of Nonribosomal Peptide Synthetases (Nrps), Fmoa3 (S1046A)-Amppnp Bound Form
Organism: Streptomyces sp. sp080513ge-23
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2021-02-24 Classification: BIOSYNTHETIC PROTEIN Ligands: ANP |
 |
Organism: Sphingomonas sp. a1
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2014-09-24 Classification: OXIDOREDUCTASE |
 |
Organism: Sphingomonas sp. a1
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2014-09-24 Classification: OXIDOREDUCTASE |
 |
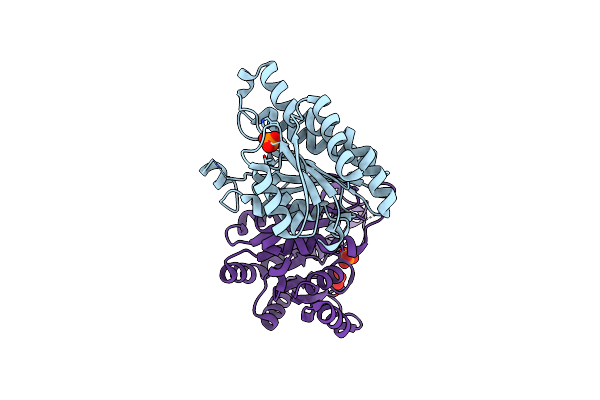
Organism: Human immunodeficiency virus type 1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-06-25 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 4BI, CD |
 |
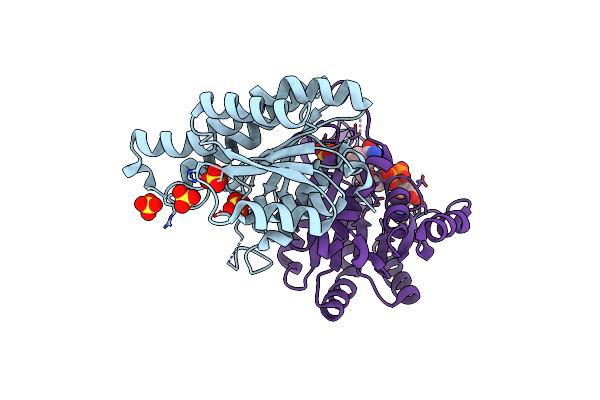
Crystal Structure Of Nadh-Dependent Reductase A1-R' Responsible For Alginate Metabolism
Organism: Sphingomonas sp. a1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-06-25 Classification: OXIDOREDUCTASE Ligands: PO4 |
 |
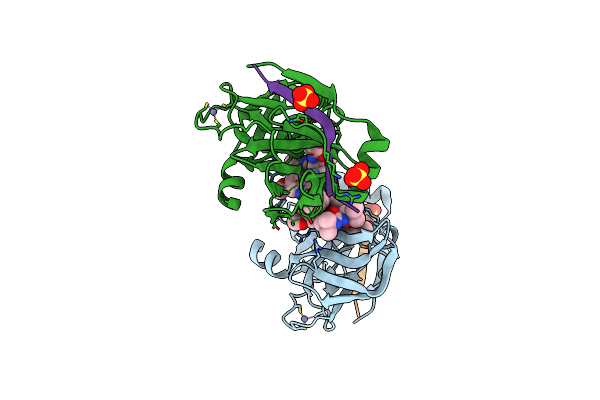
Organism: Sphingomonas sp. a1
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2014-06-25 Classification: OXIDOREDUCTASE Ligands: SO4, NAD |
 |
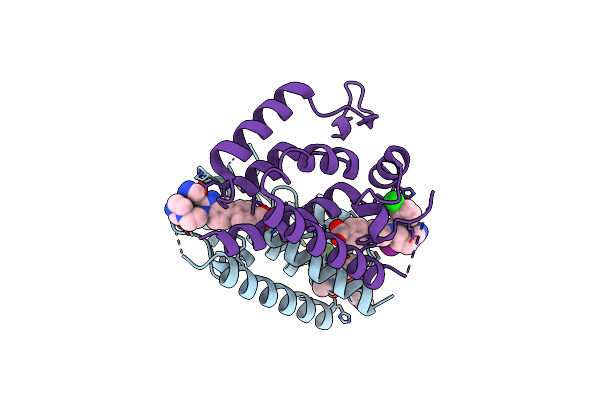
Organism: Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-03-26 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, 1RR, SO4 |
 |
Crystal Structure Of The N-Terminal Domain Of Hiv-1 Capsid In Complex With Inhibitor Bd3
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-04-25 Classification: STRUCTURAL PROTEIN/INHIBITOR Ligands: 0OE, 0OF, IOD |
 |
Crystal Structure Of The N-Terminal Domain Of Hiv-1 Capsid In Complex With Inhibitor Bm4
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-04-25 Classification: STRUCTURAL PROTEIN/INHIBITOR Ligands: 0OE, 0OG |
 |
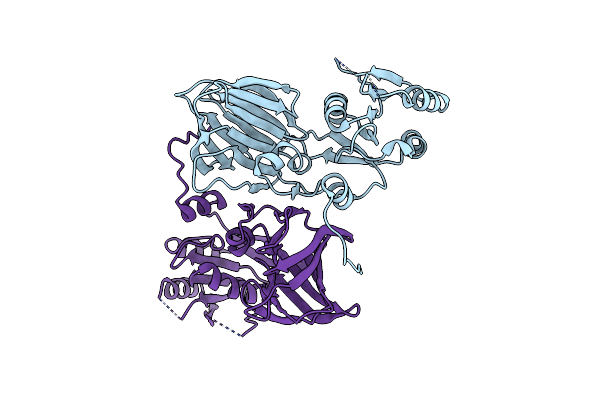
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-03-16 Classification: TRANSFERASE Ligands: NAI, MPD |
 |
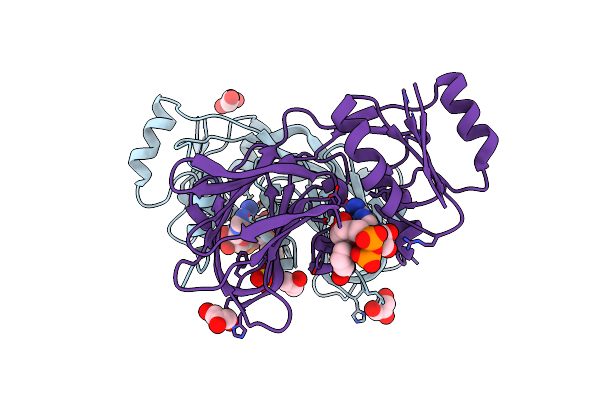
Crystal Structure Of Inorganic Polyphosphate/Atp-Nad Kinase From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2005-01-18 Classification: TRANSFERASE |
 |
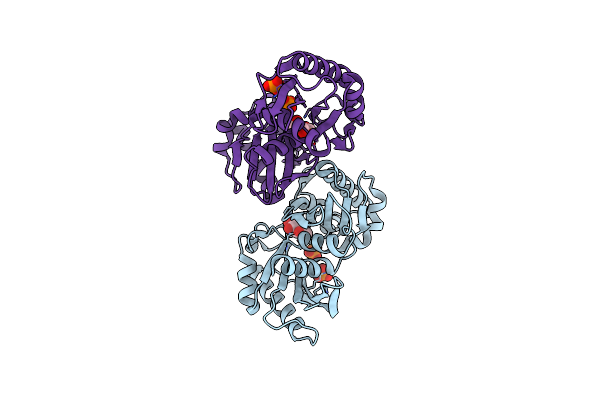
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2005-01-18 Classification: TRANSFERASE Ligands: NAD, GOL, SO4 |
 |
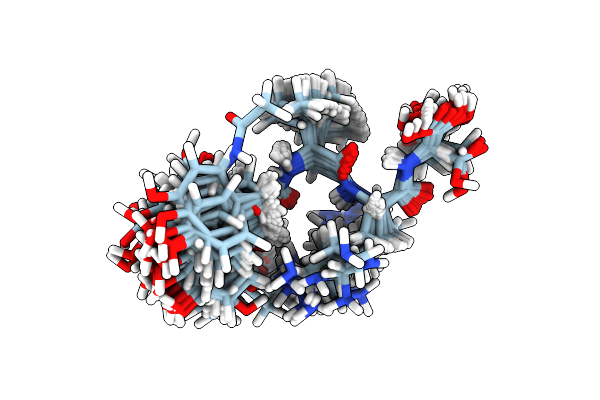
Crystal Structure Of Inorganic Polyphosphate/Atp-Glucomannokinase From Arthrobacter Sp. Strain Km At 1.8 A Resolution
Organism: Arthrobacter sp.
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2004-09-28 Classification: TRANSFERASE Ligands: BGC, PO4 |
 |
Bound Conformation Of N-Terminal Cleavage Product Peptide Mimic (P1-P9 Of Release Site) While Bound To Hcmv Protease As Determined By Transferred Noesy Experiments (P1-P5 Shown Only), Nmr, 32 Structures
|