Search Count: 15
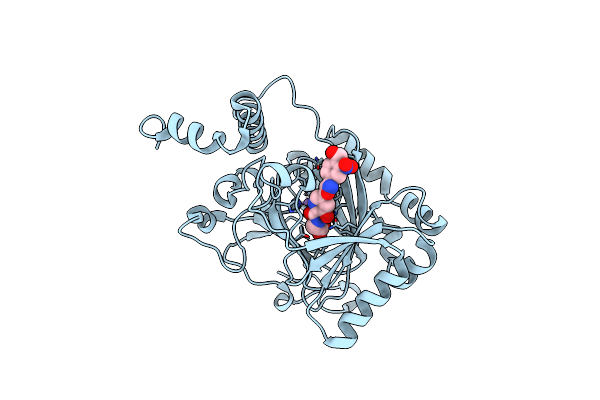 |
Factor Inhibiting Hif (Fih) In Complex With Manganese And 3-Hydroxy-5-(3-(4-(Hydroxymethyl)-3-Nitrophenyl)Isoxazol-5-Yl)Picolinoyl)Glycine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: OXIDOREDUCTASE Ligands: MN, A1IZ5 |
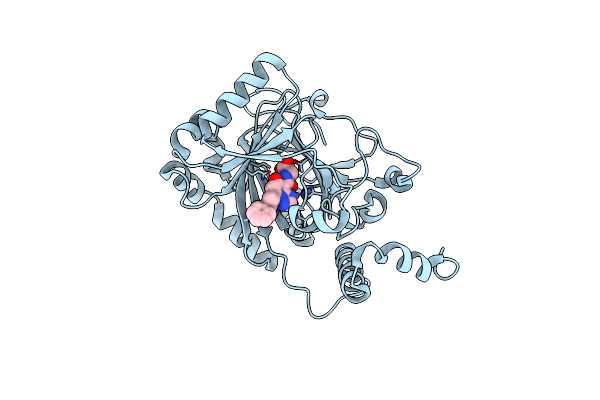 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: MN, A1IF7 |
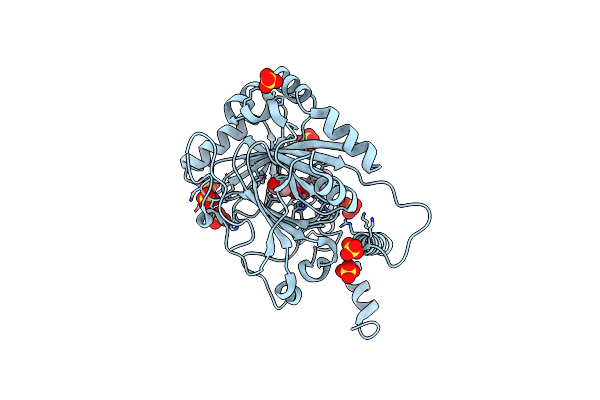 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: SO4, A1L2L, ZN |
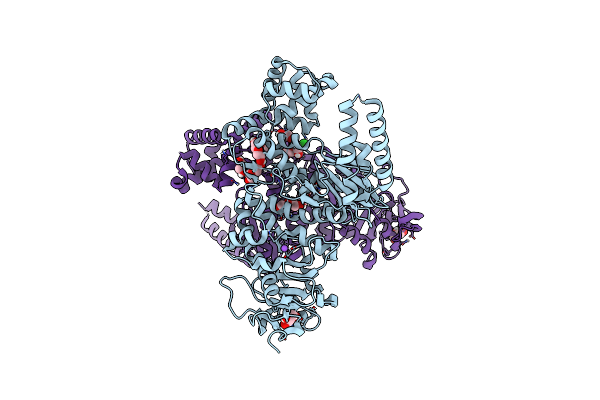 |
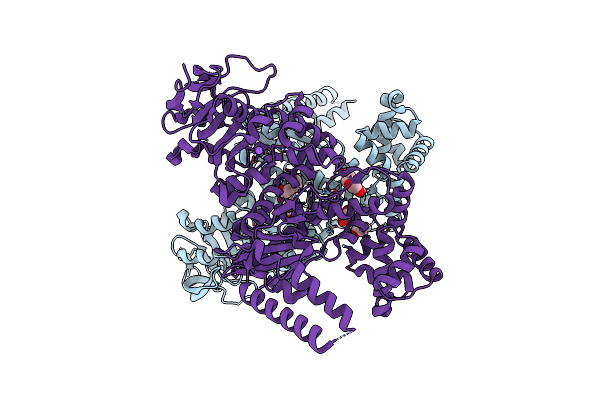
Plasmodium Falciparum Apicoplast Dna Polymerase (Exo-Minus) Without Affinity Tag
Organism: Plasmodium falciparum (isolate 3d7)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-10-26 Classification: REPLICATION, Transferase Ligands: GOL, NA, CL, PEG, EDO |
 |
Plasmodium Falciparum Apicoplast Dna Polymerase (Exo-Minus) Without Affinity Tag
Organism: Plasmodium falciparum (isolate 3d7)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-10-19 Classification: REPLICATION, TRANSFERASE Ligands: PEG, CL, NA, EDO |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-03-30 Classification: OXIDOREDUCTASE/RNA Ligands: GOL, MN, SO4 |
 |
Crystal Structure Of Human Alkbh5 In Complex With 2-Oxoglutarate (2Og) And M6A-Containing Ssrna
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-03-30 Classification: OXIDOREDUCTASE/RNA Ligands: MN, AKG |
 |
Crystal Structure Of Human Alkbh5 In Complex With N-Oxalylglycine (Nog) And M6A-Containing Ssrna
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-03-30 Classification: OXIDOREDUCTASE/RNA Ligands: MN, OGA, FMT |
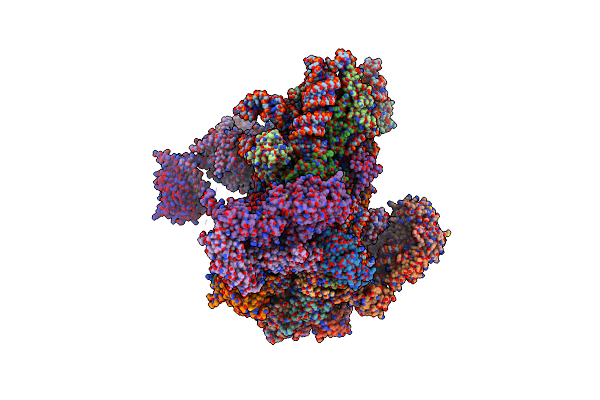 |
Structure Of A Partial Yeast 48S Preinitiation Complex With Eif5 N-Terminal Domain (Model C1)
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Saccharomyces cerevisiae, Kluyveromyces lactis (strain atcc 8585 / cbs 2359 / dsm 70799 / nbrc 1267 / nrrl y-1140 / wm37)
Method: ELECTRON MICROSCOPY Release Date: 2018-12-05 Classification: RIBOSOME Ligands: MG, ZN, MET, GCP |
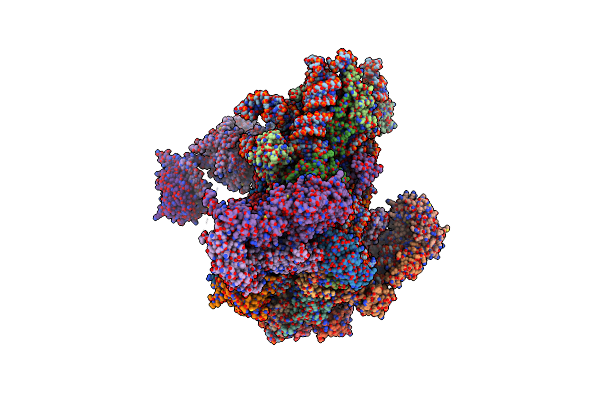 |
Structure Of A Partial Yeast 48S Preinitiation Complex With Eif5 N-Terminal Domain (Model C2)
Organism: Saccharomyces cerevisiae, Kluyveromyces lactis (strain atcc 8585 / cbs 2359 / dsm 70799 / nbrc 1267 / nrrl y-1140 / wm37)
Method: ELECTRON MICROSCOPY Resolution:3.02 Å Release Date: 2018-12-05 Classification: RIBOSOME Ligands: MG, ZN, MET, GCP |
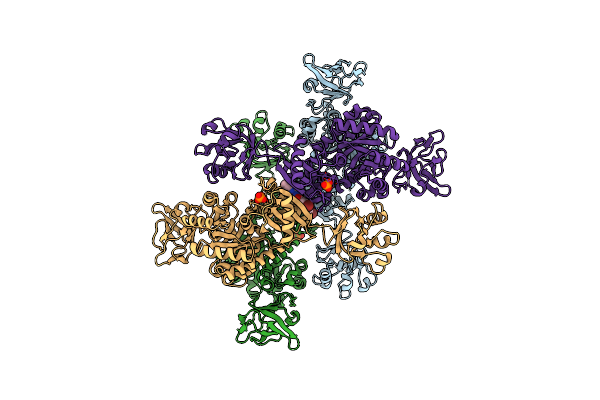 |
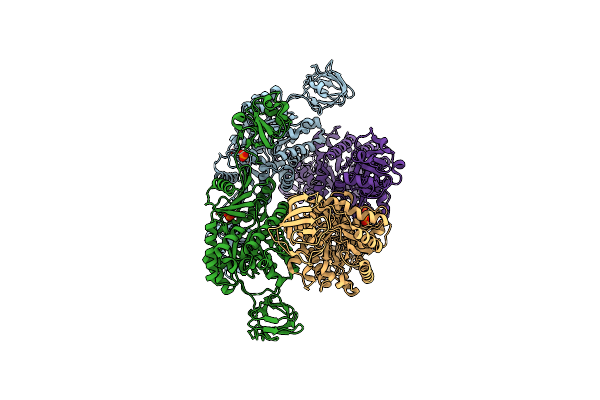
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2012-06-06 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: I30, PO4 |
 |
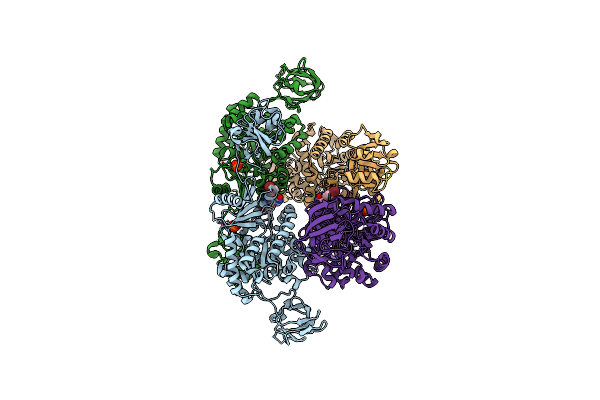
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2011-10-26 Classification: TRANSFERASE Ligands: PO4 |
 |
Crystal Structure Of S. Aureus Pyruvate Kinase In Complex With A Naturally Occurring Bis-Indole Alkaloid
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2011-10-26 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: PO4, 09C |
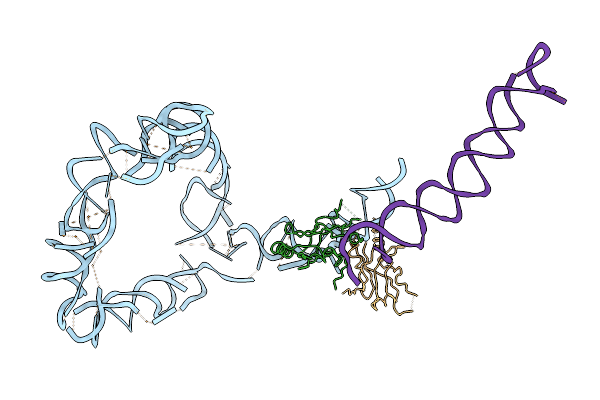 |
Organism: Thermus thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2007-01-23 Classification: RNA BINDING PROTEIN/RNA |
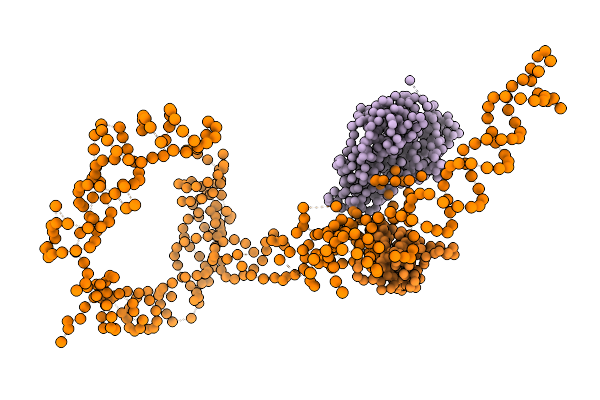 |
Coordinates Of Tmrna, Smpb, Ef-Tu And H44 Fitted Into Cryo-Em Map Of The 70S Ribosome And Tmrna Complex
Organism: Thermus thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2005-04-19 Classification: RNA binding protein/RNA |

