Search Count: 458
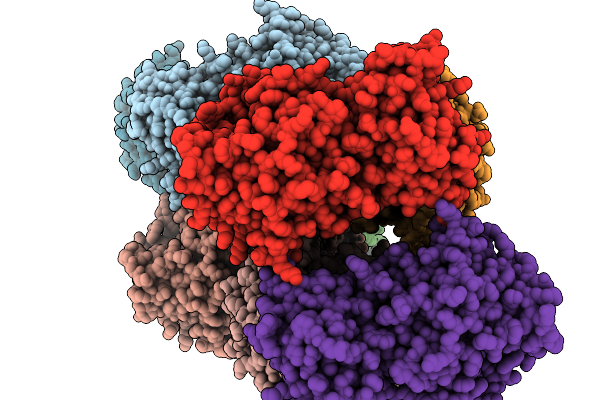 |
Crystal Structure Of Pseudopedobacter Saltans Gh43 Beta-Xylosidase In Complex With Xylose.
Organism: Pseudopedobacter saltans dsm 12145
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: HYDROLASE Ligands: CA, XLS |
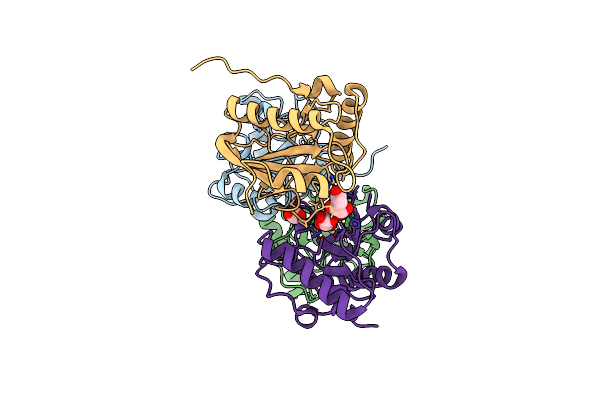 |
Crystal Structure Of The Complex Of Camel Peptidoglycan Recognition Protein, Pgrp-S With Malic Acid And Oxalic Acid At 2.3 A Resolution
Organism: Camelus dromedarius
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2025-05-14 Classification: IMMUNE SYSTEM Ligands: MLT, OXD |
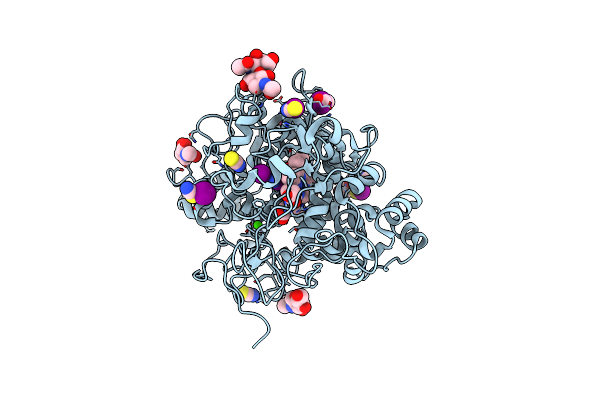 |
Structure Of The Intermediate Of Lactoperoxidase Formed With Thiocynate And Hydrogen Peroxidase At 1.99 A Resolution.
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-03-26 Classification: OXIDOREDUCTASE Ligands: HEM, CA, NAG, IOD, SCN, PEO, GOL |
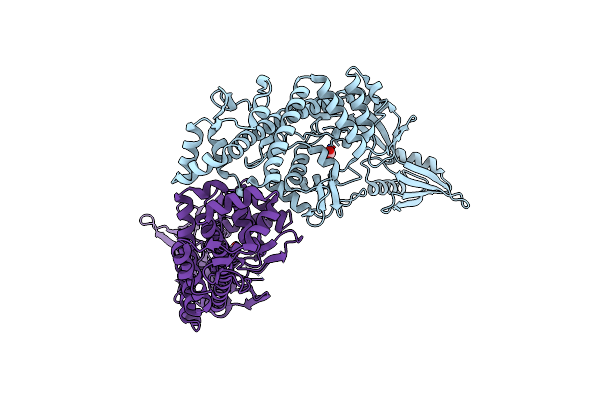 |
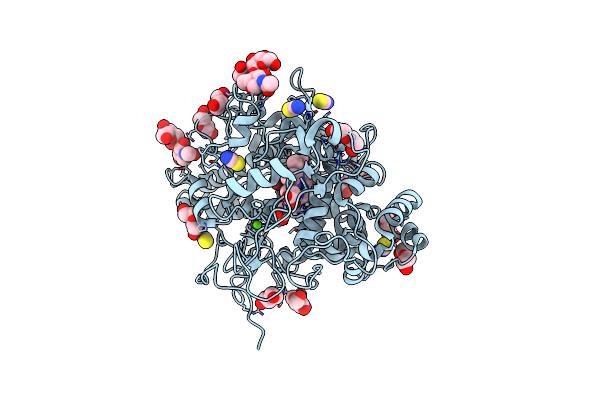
Organism: Salmonella enterica subsp. enterica serovar typhi str. ct18
Method: X-RAY DIFFRACTION Release Date: 2024-05-08 Classification: ISOMERASE Ligands: CO3 |
 |
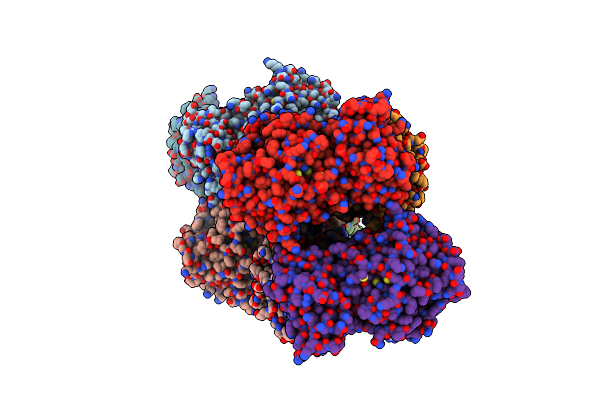
Crystal Structure Of The Complex Of Lactoperoxidase With Four Inorganic Substrates, Scn, I, Br And Cl
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-03-13 Classification: OXIDOREDUCTASE Ligands: HEM, CA, IOD, BR, CL, PEG, SCN, NAG |
 |
Structure Of Gh43 Family Enzyme, Xylan 1, 4 Beta- Xylosidase From Pseudopedobacter Saltans
Organism: Pseudopedobacter saltans dsm 12145
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2023-11-15 Classification: HYDROLASE Ligands: CA |
 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-08-09 Classification: DNA BINDING PROTEIN/DNA Ligands: EDO, MPD, MES |
 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2023-08-09 Classification: DNA BINDING PROTEIN/DNA Ligands: GOL, EDO, MPD, NA |
 |
Structure Of The Ternary Complex Of Lactoperoxidase With Substrate Nitric Oxide (No) And Product Nitrite Ion (No2) At 1.98 A Resolution
Organism: Capra hircus
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2023-04-05 Classification: OXIDOREDUCTASE Ligands: NAG, NO, NO2, NO3, SCN, EDO, IOD, HEM, CA, OSM, NA |
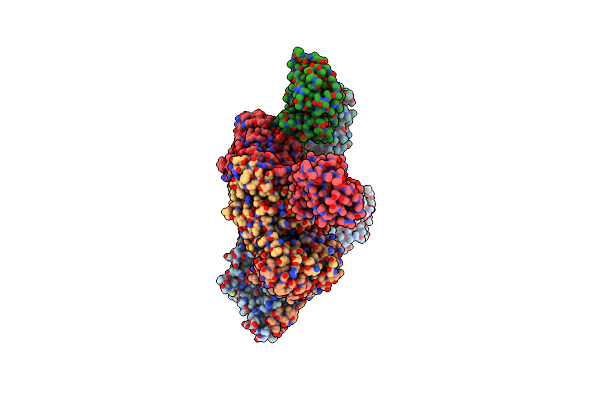 |
Organism: Lates calcarifer, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-11-02 Classification: REPLICATION/DNA Ligands: ATP, MG |
 |
Crystal Structure Of The Complex Of Proliferating Cell Nuclear Antigen (Pcna) From Leishmania Donovani With 1,5-Bis (4-Amidinophenoxy) Pentane (Pnt) At 2.95 A Resolution
Organism: Leishmania donovani
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2022-06-15 Classification: DNA BINDING PROTEIN Ligands: PNT |
 |
Structure Of The Complex Of Camel Peptidoglycan Recognition Protein-Short (Pgrp-S) With Heptanoic Acid At 2.15 A Resolution.
Organism: Camelus dromedarius
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2022-06-15 Classification: IMMUNE SYSTEM Ligands: EDO, PEO, CO3, NA, SHV, MPD |
 |
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2022-05-18 Classification: SUGAR BINDING PROTEIN Ligands: SO4 |
 |
Crystal Structure Lpqy In Complex With Trehalose From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2022-05-18 Classification: SUGAR BINDING PROTEIN Ligands: SO4 |
 |
Crystal Structure Of The Ternary Complex Of Peptidoglycan Recognition Protein, Pgrp-S With Hexanoic And Tartaric Acids At 2.07 A Resolution.
Organism: Camelus dromedarius
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2022-05-11 Classification: IMMUNE SYSTEM Ligands: TLA, EDO, ACT, CL, 6NA, GOL |
 |
Crystal Structure Of P73 Dna Binding Domain Complex Bound With 1 Bp And 2 Bp Spacer Dna Response Elements.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2022-04-13 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
 |
Crystal Structure Of Bovine Lactoperoxidase With Hydrogen Peroxide Trapped Between Heme Iron And His109 At 1.69 A Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-12-30 Classification: OXIDOREDUCTASE Ligands: HEM, CA, NAG, IOD, GOL, EDO, ZN, OSM, PEO |
 |
Crystal Structure Of Ternary Complexes Of Lactoperoxidase With Hydrogen Peroxide At 1.70 A Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-12-30 Classification: OXIDOREDUCTASE Ligands: HEM, CA, NAG, IOD, EDO, ZN, OSM, PEO |
 |
Crystal Structure Of The Complex Of Lactoperoxidase With Hydrogen Peroxide At 1.77A Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2020-12-16 Classification: OXIDOREDUCTASE Ligands: HEM, CA, NAG, IOD, GOL, EDO, ZN, OSM, PEO |
 |
Crystal Structure Of Potassium Induced Heme Modification In Yak Lactoperoxidase At 2.20 A Resolution
Organism: Bos mutus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-12-16 Classification: OXIDOREDUCTASE Ligands: NAG, CA, HEM, ZN, CL, K |

