Search Count: 22
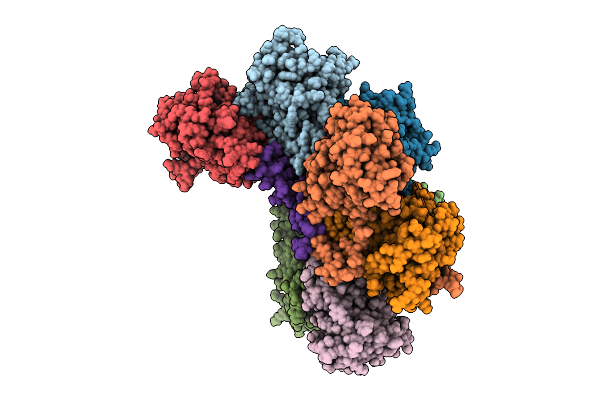 |
Proteasome Core Particle Assembly Intermediate 5-Alpha/3-Beta/Ump1 Purified From Saccharomyces Cerevisiae.
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: HYDROLASE |
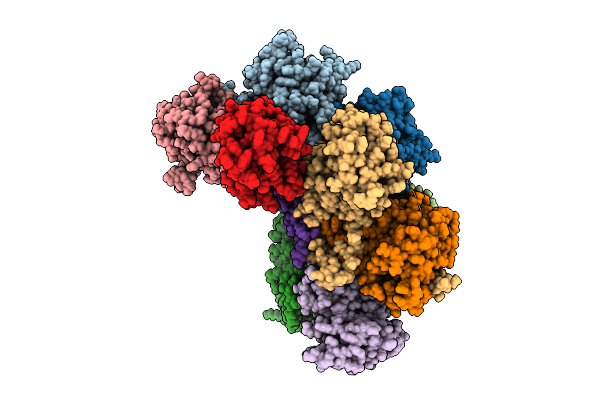 |
Proteasome Core Particle Assembly Intermediate 5-Alpha/4-Beta/Ump1 Purified From Saccharomyces Cerevisiae.
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: HYDROLASE |
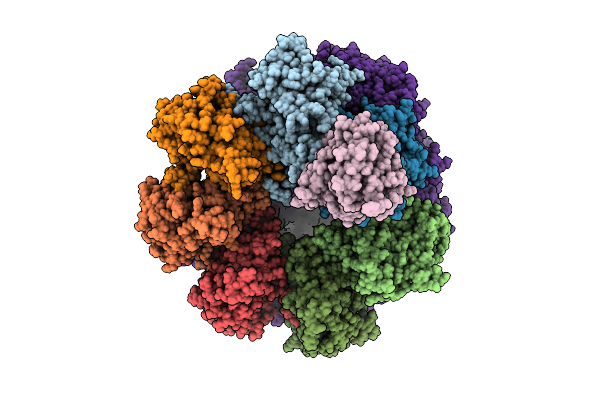 |
Proteasome Core Particle Assembly Intermediate Blm10:Alpha-Ring Purified From Saccharomyces Cerevisiae.
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: HYDROLASE |
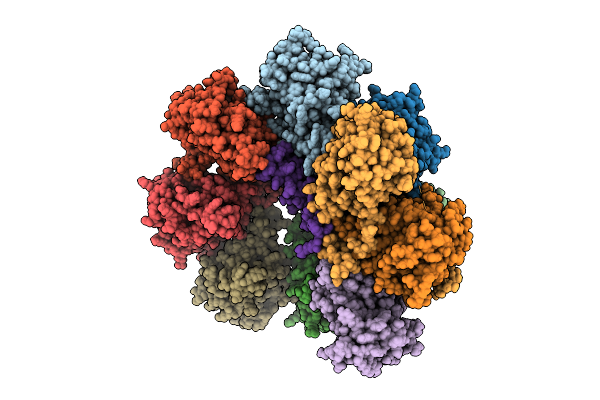 |
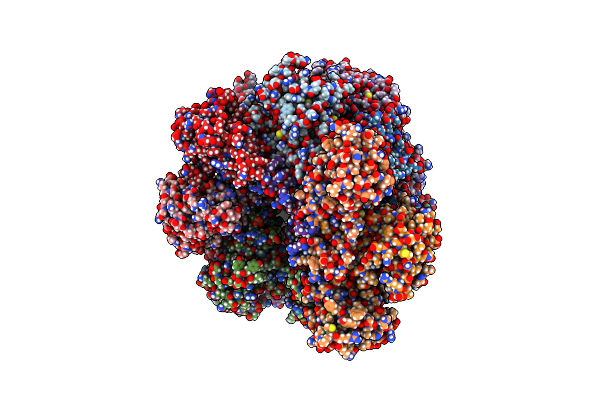
Core Particle Assembly Intermediate Capless 13S Purified From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: HYDROLASE |
 |
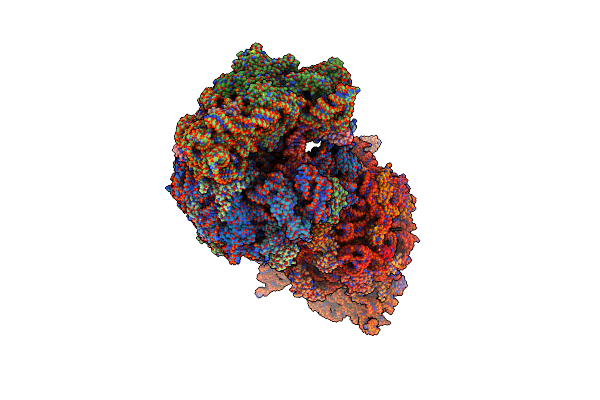
Proteasome Core Particle Assembly Intermediate Blm10:13S Purified From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-12-04 Classification: HYDROLASE |
 |
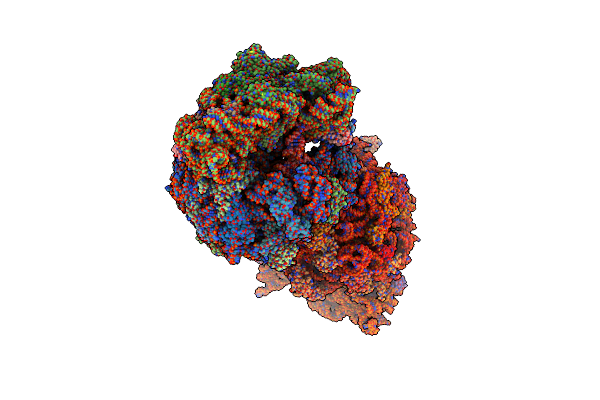
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Lasso Peptide Lariocidin, Mrna, Aminoacylated A-Site Phe-Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.50A Resolution
Organism: Escherichia coli, Paenibacillus, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-11-20 Classification: RIBOSOME Ligands: MG, K, ZN, SF4 |
 |
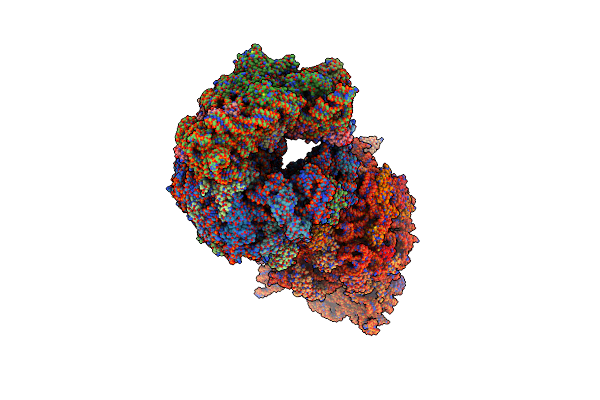
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Lasso Peptide Lariocidin B, Mrna, Aminoacylated A-Site Phe-Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.60A Resolution
Organism: Escherichia coli, Paenibacillus, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-11-20 Classification: RIBOSOME Ligands: MG, K, ZN, SF4 |
 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Lasso Peptide Lariocidin And Protein Y At 2.60A Resolution
Organism: Escherichia coli k-12, Paenibacillus, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-11-20 Classification: RIBOSOME Ligands: MG, ARG, MPD, ZN, SF4 |
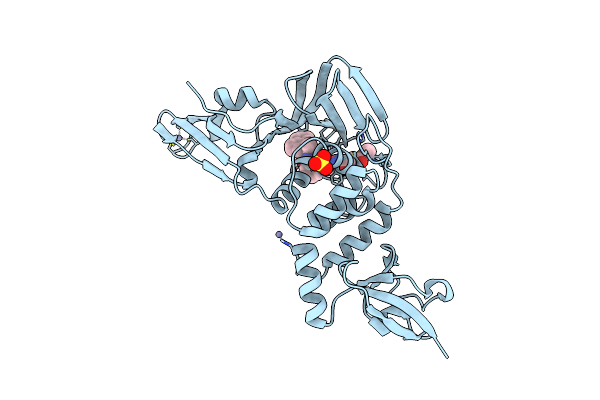 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2023-04-05 Classification: HYDROLASE/INHIBITOR Ligands: WUK, ZN, SO4 |
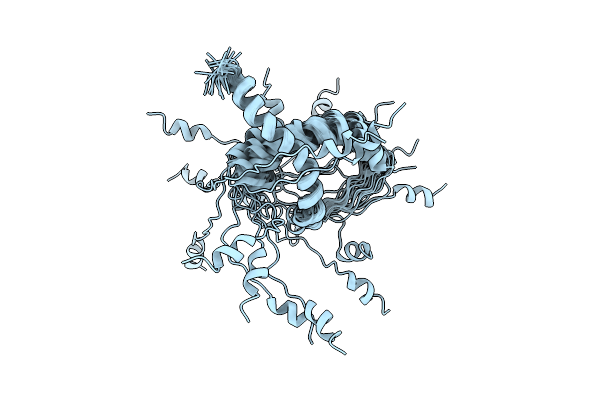 |
Organism: Human immunodeficiency virus 1
Method: SOLUTION NMR Release Date: 2022-10-26 Classification: ANTIVIRAL PROTEIN |
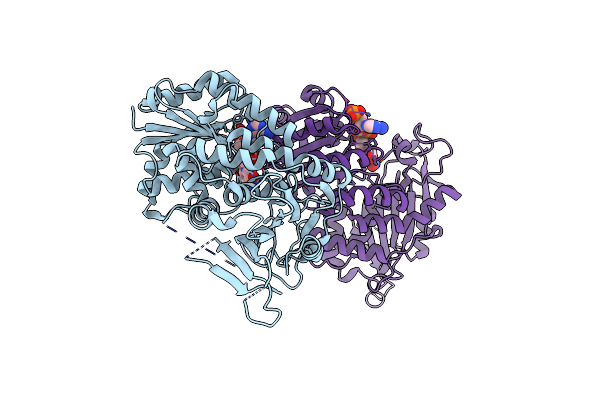 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2022-08-17 Classification: OXIDOREDUCTASE Ligands: NAP |
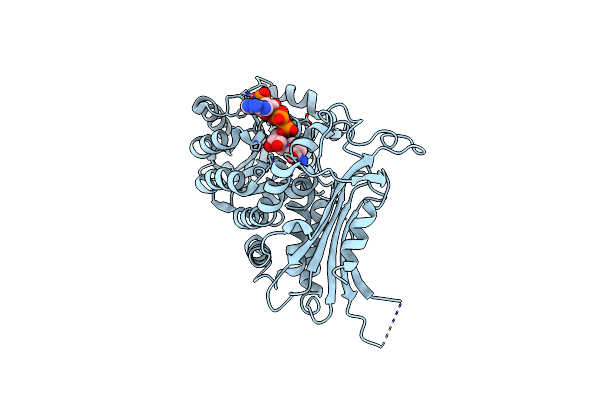 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.65 Å Release Date: 2022-08-17 Classification: OXIDOREDUCTASE Ligands: NAP |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-04-14 Classification: HYDROLASE |
 |
Cryo-Em Structure Of Pre-15S Proteasome Core Particle Assembly Intermediate Purified From Pre3-1 Proteasome Mutant (G34D)
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-04-14 Classification: HYDROLASE |
 |
Cryo-Em Structure Of 13S Proteasome Core Particle Assembly Intermediate Purified From Pre3-1 Proteasome Mutant (G34D)
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-04-14 Classification: HYDROLASE |
 |
Organism: Kitasatospora setae (strain atcc 33774 / dsm 43861 / jcm 3304 / kcc a-0304 / nbrc 14216 / km-6054)
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2020-03-18 Classification: OXIDOREDUCTASE Ligands: NAP, VES, 2EC, YAS, NDP |
 |
Organism: Kitasatospora setae (strain atcc 33774 / dsm 43861 / jcm 3304 / kcc a-0304 / nbrc 14216 / km-6054)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-12-11 Classification: OXIDOREDUCTASE Ligands: VES, CL |
 |
Organism: Kitasatospora setae (strain atcc 33774 / dsm 43861 / jcm 3304 / kcc a-0304 / nbrc 14216 / km-6054)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-12-11 Classification: OXIDOREDUCTASE Ligands: NDP |
 |
Serial Femtosecond X-Ray Crystallography Structure Of Ecr In Complex With Nadph
Organism: Kitasatospora setae (strain atcc 33774 / dsm 43861 / jcm 3304 / kcc a-0304 / nbrc 14216 / km-6054)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-12-11 Classification: OXIDOREDUCTASE Ligands: NDP |
 |
X-Ray Structure Of A C-3'-Methyltransferase In Complex With S-Adenosylmethionine And Dtmp
Organism: Micromonospora chalcea
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-06-30 Classification: TRANSFERASE Ligands: SAM, ZN, TMP, PO4 |

