Search Count: 17
 |
Formation Of Left-Handed Helices By C2'-Fluorinated Nucleic Acids Under Physiological Salt Conditions
|
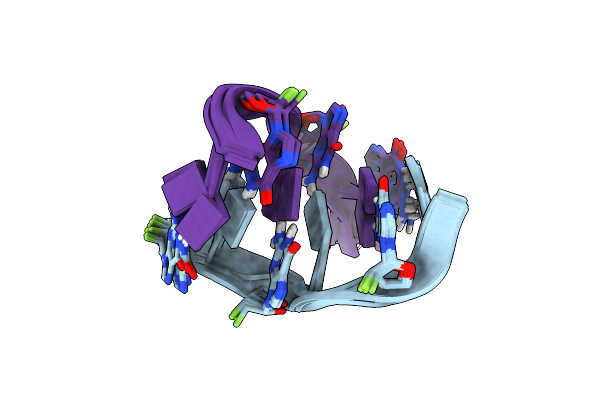 |
Formation Of Left-Handed Helices By C2'-Fluorinated Nucleic Acids Under Physiological Salt Conditions
|
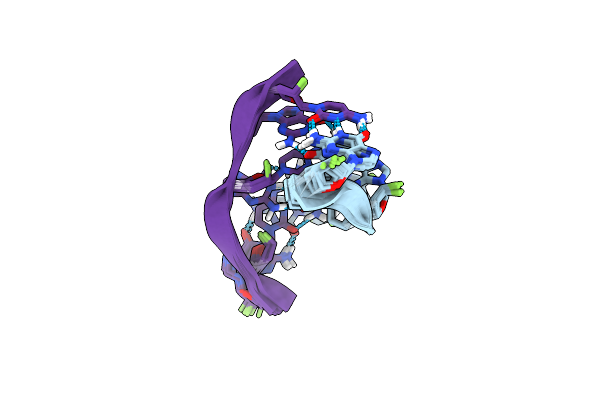 |
Formation Of Left-Handed Helices By C2'-Fluorinated Nucleic Acids Under Physiological Salt Conditions
|
 |
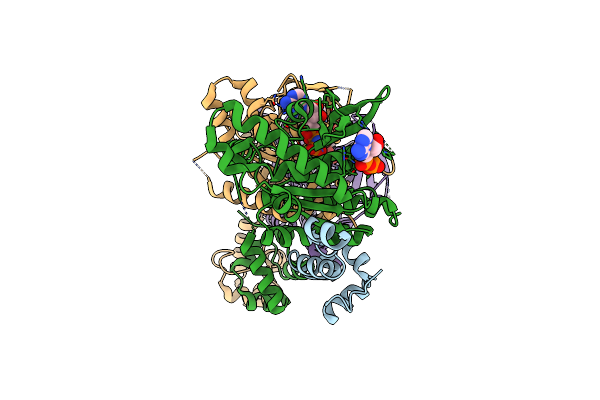
Crystal Structure Of Haloacid Dehalogenase-Like Hydrolase Family Enzyme From Staphylococcus Lugdunensis
Organism: Staphylococcus lugdunensis
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2023-12-27 Classification: HYDROLASE Ligands: PEG, EDO, OXM, FMT |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-06-21 Classification: RECOMBINATION Ligands: ADP, ANP, MG |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-06-21 Classification: RECOMBINATION Ligands: ANP |
 |
Crystal Structure Of E.Coli Bama Beta-Barrel In Complex With Darobactin (Crystal Form 1)
Organism: Escherichia coli o157:h7, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-04-21 Classification: MEMBRANE PROTEIN Ligands: C8E, MG |
 |
Crystal Structure Of E.Coli Bama Beta-Barrel In Complex With Darobactin (Crystal Form 2)
Organism: Escherichia coli o157:h7, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-04-21 Classification: MEMBRANE PROTEIN Ligands: MG, C8E |
 |
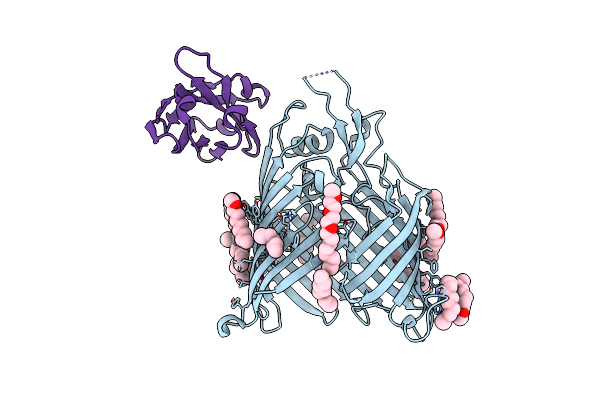
Organism: Escherichia coli (strain k12), Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-04-21 Classification: MEMBRANE PROTEIN |
 |
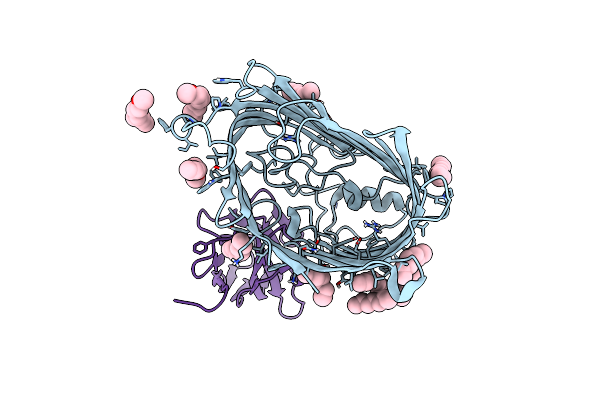
Organism: Escherichia coli o157:h7, Lama glama
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2019-06-26 Classification: MEMBRANE PROTEIN Ligands: C8E |
 |
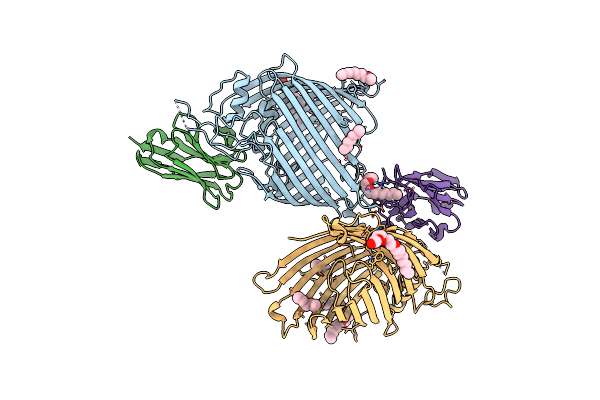
Organism: Escherichia coli o157:h7, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-06-26 Classification: MEMBRANE PROTEIN Ligands: C8E |
 |
Organism: Escherichia coli o157:h7, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2019-06-26 Classification: MEMBRANE PROTEIN Ligands: C8E |
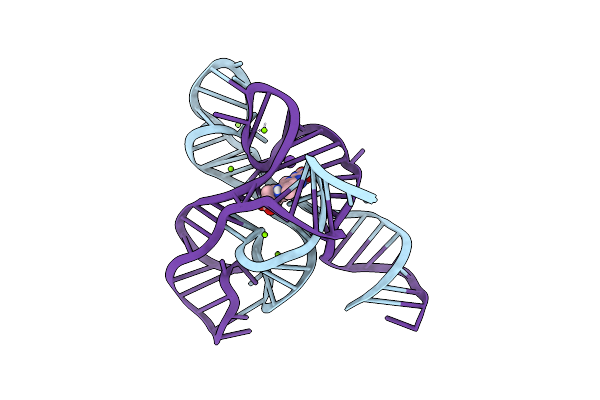 |
Organism: Fusobacterium nucleatum
Method: X-RAY DIFFRACTION Resolution:3.03 Å Release Date: 2018-09-05 Classification: RNA Ligands: MG, GZ7, K |
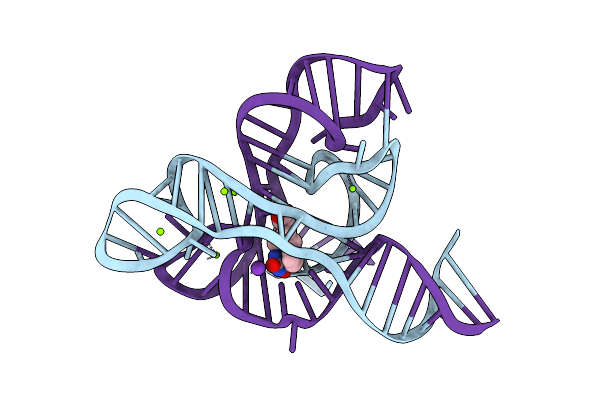 |
Organism: Fusobacterium nucleatum
Method: X-RAY DIFFRACTION Resolution:2.88 Å Release Date: 2018-09-05 Classification: RNA Ligands: MG, GZG, K |
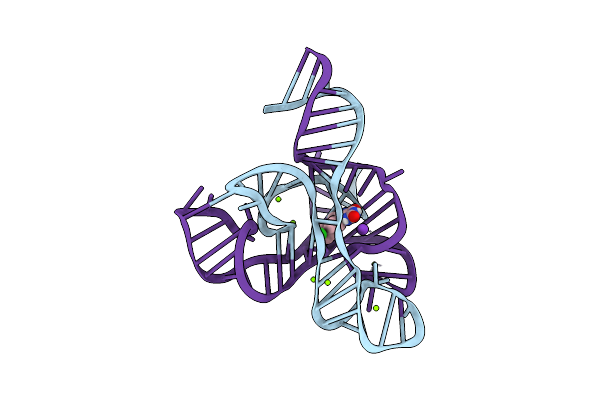 |
Organism: Fusobacterium nucleatum
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-09-05 Classification: RNA Ligands: CL, MG, GZ4, K |
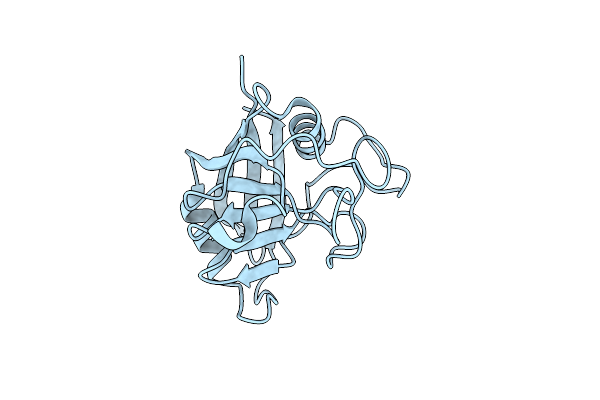 |
Organism: Triticum aestivum
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2013-03-27 Classification: ISOMERASE |
 |
Structural And Biochemical Characterization Of A Cytosolic Wheat Cyclophilin Tacypa-1
Organism: Triticum aestivum, Tolypocladium inflatum
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2013-03-27 Classification: ISOMERASE/INHIBITOR |

